Iteration
Lecture 13
Dr. Benjamin Soltoff
Cornell University
INFO 2950 - Spring 2024
March 7, 2024
Announcements
Announcements
- Lab 04
- Homework 04
- Project proposals
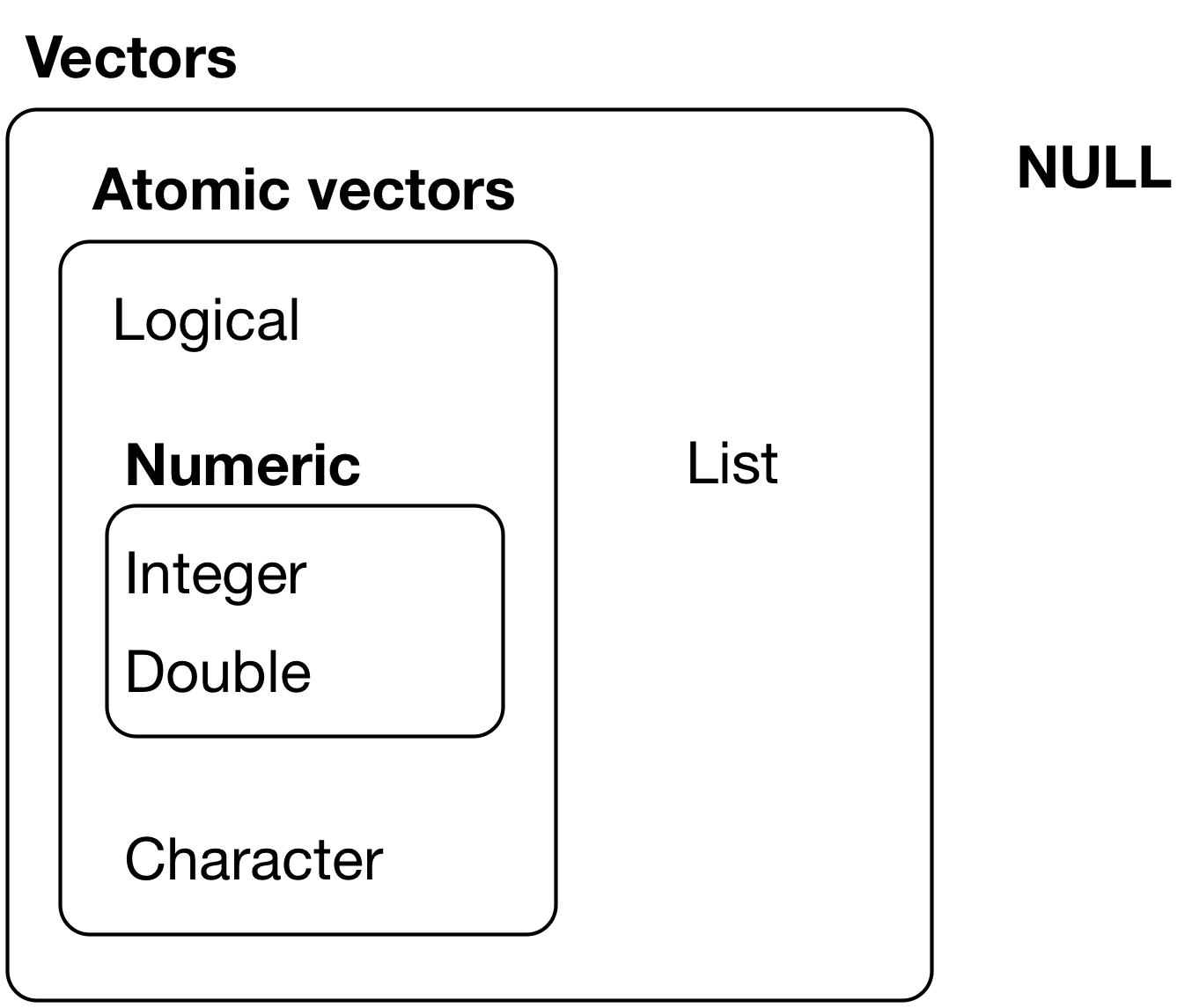
Atomic vectors
Subsetting vectors with [] and [[]]
- With positive integers
- With negative integers
- Don’t mix positive and negative
Subset with a logical vector
Lists
Lists
Lists: str()
Store a mix of objects

Nested lists
Stealth lists
tibble [53,940 × 10] (S3: tbl_df/tbl/data.frame)
$ carat : num [1:53940] 0.23 0.21 0.23 0.29 0.31 0.24 0.24 0.26 0.22 0.23 ...
$ cut : Ord.factor w/ 5 levels "Fair"<"Good"<..: 5 4 2 4 2 3 3 3 1 3 ...
$ color : Ord.factor w/ 7 levels "D"<"E"<"F"<"G"<..: 2 2 2 6 7 7 6 5 2 5 ...
$ clarity: Ord.factor w/ 8 levels "I1"<"SI2"<"SI1"<..: 2 3 5 4 2 6 7 3 4 5 ...
$ depth : num [1:53940] 61.5 59.8 56.9 62.4 63.3 62.8 62.3 61.9 65.1 59.4 ...
$ table : num [1:53940] 55 61 65 58 58 57 57 55 61 61 ...
$ price : int [1:53940] 326 326 327 334 335 336 336 337 337 338 ...
$ x : num [1:53940] 3.95 3.89 4.05 4.2 4.34 3.94 3.95 4.07 3.87 4 ...
$ y : num [1:53940] 3.98 3.84 4.07 4.23 4.35 3.96 3.98 4.11 3.78 4.05 ...
$ z : num [1:53940] 2.43 2.31 2.31 2.63 2.75 2.48 2.47 2.53 2.49 2.39 ...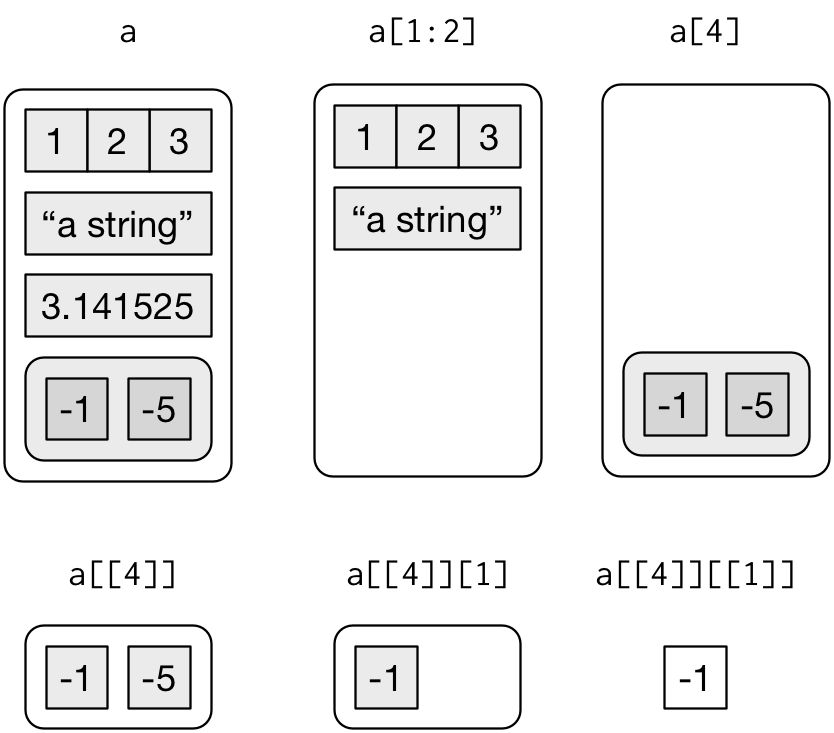
Iteration
Iteration
Iteration three ways
forloopsmap_*()functionsacross()
Iteration with for loops
Iteration with for loop
Output
Sequence
Body
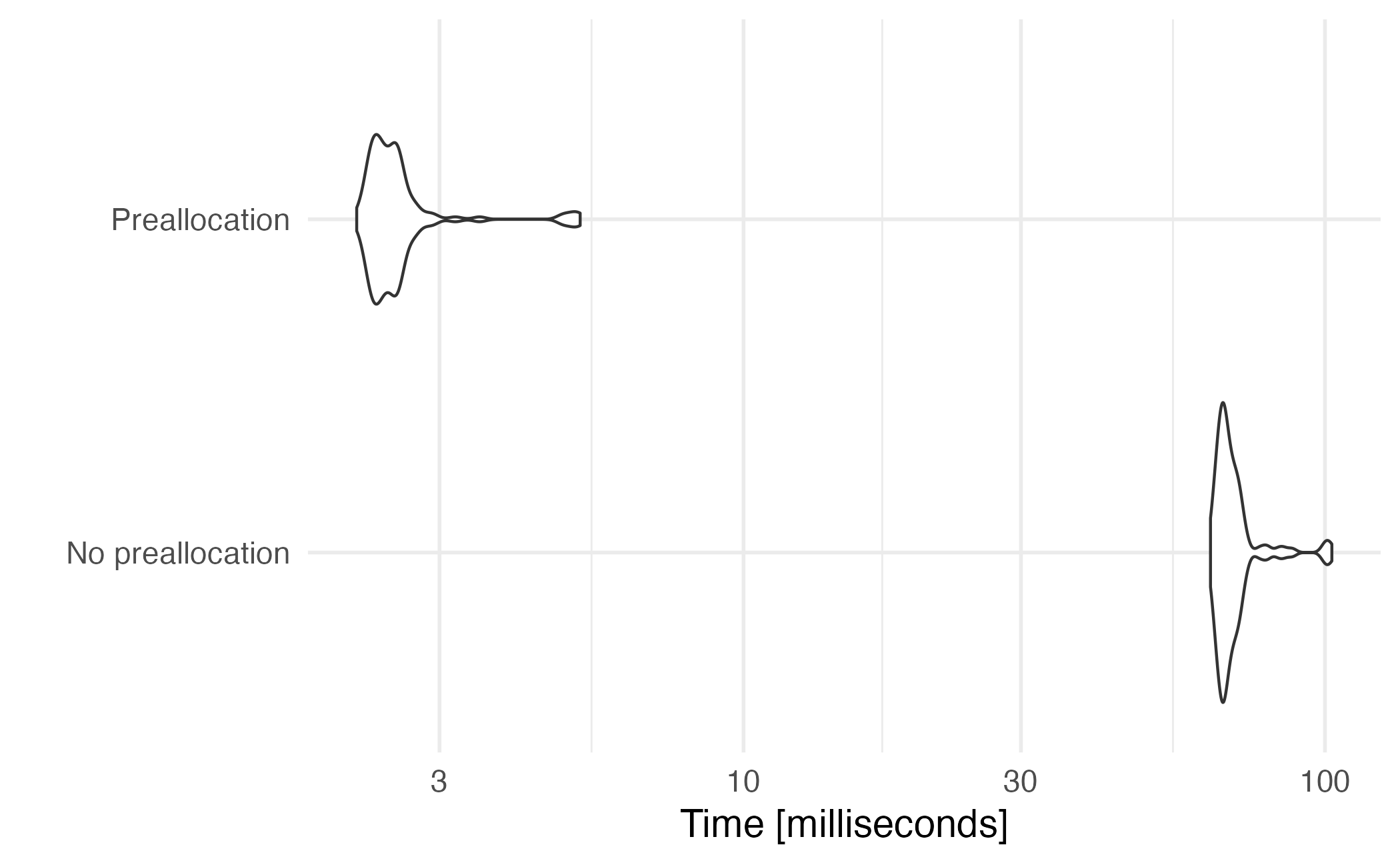
Preallocation
Iteration with map_*() functions
Map functions
- Why
forloops are good - Why
map()functions may be better - Types of
map()functionsmap()makes a listmap_lgl()makes a logical vectormap_int()makes an integer vectormap_dbl()makes a double vectormap_chr()makes a character vector
Map functions
Map functions
a b c d
0.07462564 0.20862196 -0.42455887 0.32204455 Application exercise
ae-11
- Go to the course GitHub org and find your
ae-11(repo name will be suffixed with your GitHub name). - Clone the repo in RStudio Workbench, open the R script in the repo, and follow along and complete the exercises.
- Render, commit, and push your edits by the AE deadline – end of tomorrow
Recap
- Use
[],[[]], and$notation to extract elements from an atomic vector or list object forloops +map()functions are common methods for iteration in R- When using
forloops, always preallocate the output vector map()functions are a family of functions that apply a function to each element of a vector or list