hotels <- read_csv("data/hotels.csv") |>
mutate(across(where(is.character), as.factor))
count(hotels, children)# A tibble: 2 × 2
children n
<fct> <int>
1 children 4039
2 none 45961Lecture 23
Cornell University
INFO 2950 - Spring 2024
April 18, 2024
ae-21ae-21 (repo name will be suffixed with your GitHub name).recipesrecipesrecipe()recipe()Creates a recipe for a set of variables
step_*()Adds a single transformation to a recipe. Transformations are replayed in order when the recipe is run on data.
# A tibble: 45,000 × 1
arrival_date
<date>
1 2016-04-28
2 2016-12-29
3 2016-10-17
4 2016-05-22
5 2016-03-02
6 2016-06-16
7 2017-02-13
8 2017-08-20
9 2017-08-22
10 2017-05-18
# ℹ 44,990 more rows# A tibble: 45,000 × 4
arrival_date arrival_date_dow arrival_date_month arrival_date_year
<date> <fct> <fct> <int>
1 2016-04-28 Thu Apr 2016
2 2016-12-29 Thu Dec 2016
3 2016-10-17 Mon Oct 2016
4 2016-05-22 Sun May 2016
5 2016-03-02 Wed Mar 2016
6 2016-06-16 Thu Jun 2016
7 2017-02-13 Mon Feb 2017
8 2017-08-20 Sun Aug 2017
9 2017-08-22 Tue Aug 2017
10 2017-05-18 Thu May 2017
# ℹ 44,990 more rowsstep_*()Complete list at: https://recipes.tidymodels.org/reference/index.html
step_holiday() + step_rm()Generate a set of indicator variables for specific holidays.
step_holiday() + step_rm()Rows: 45,000
Columns: 11
$ arrival_date_dow <fct> Thu, Thu, Mon, Sun, Wed, Thu, Mon, Sun, Tue,…
$ arrival_date_month <fct> Apr, Dec, Oct, May, Mar, Jun, Feb, Aug, Aug,…
$ arrival_date_year <int> 2016, 2016, 2016, 2016, 2016, 2016, 2017, 20…
$ arrival_date_AllSouls <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_AshWednesday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_ChristmasEve <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_Easter <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_ChristmasDay <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_GoodFriday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_NewYearsDay <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ arrival_date_PalmSunday <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…To predict the outcome of a new data point:
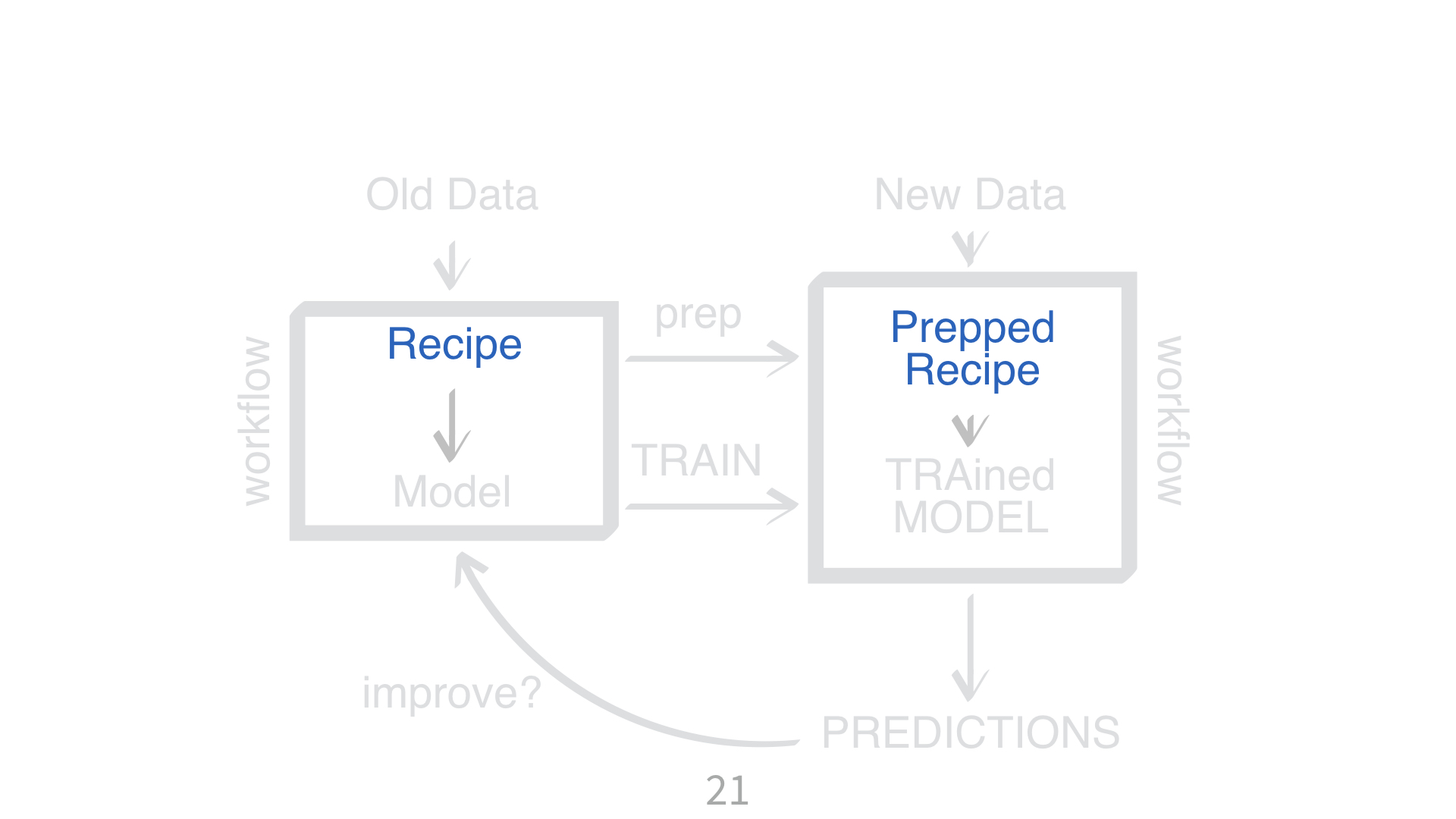
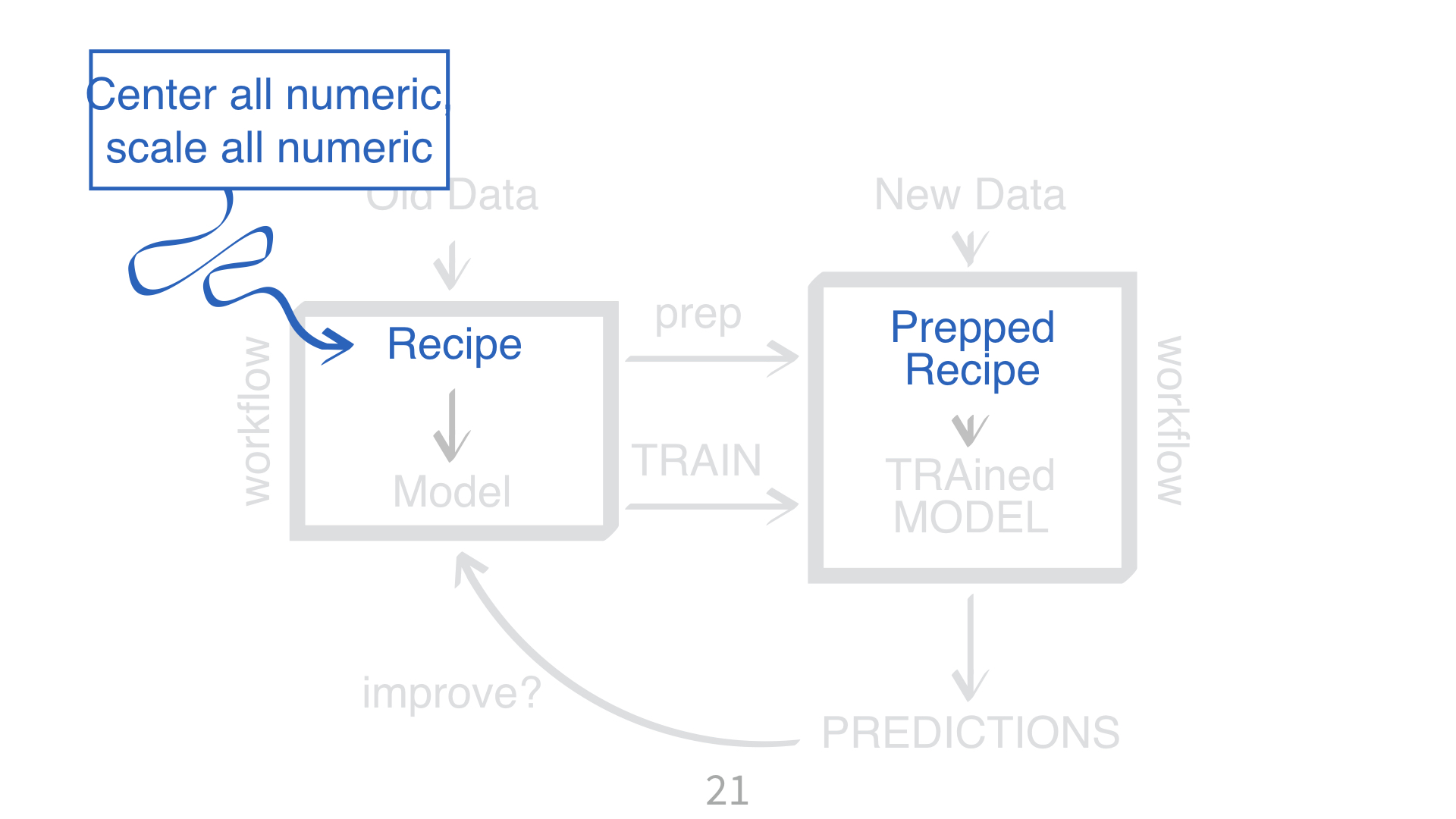
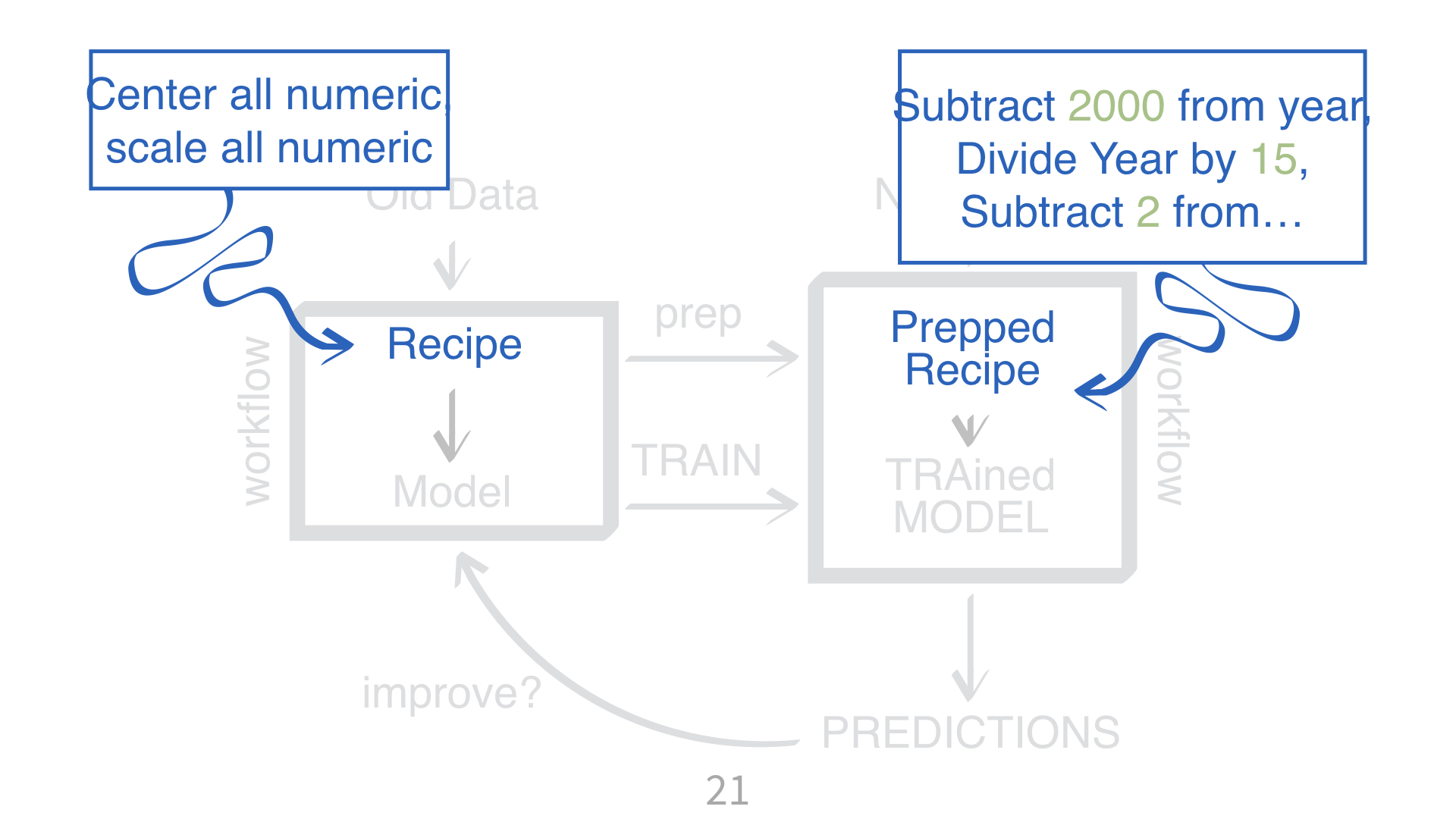
parsnipparsnipKNN requires all numeric predictors, and all need to be centered and scaled.
What does that mean?
Why do you need to “train” a recipe?
Imagine “scaling” a new data point. What do you subtract from it? What do you divide it by?
# A tibble: 5 × 1
meal
<fct>
1 SC
2 BB
3 HB
4 Undefined
5 FB # A tibble: 50,000 × 5
SC BB HB Undefined FB
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 0 0 0 0
2 0 1 0 0 0
3 0 1 0 0 0
4 0 1 0 0 0
5 0 1 0 0 0
6 0 1 0 0 0
7 0 0 1 0 0
8 0 1 0 0 0
9 0 0 1 0 0
10 1 0 0 0 0
# ℹ 49,990 more rows# A tibble: 5 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 2.38 0.0183 130. 0
2 mealFB -1.15 0.165 -6.98 2.88e-12
3 mealHB -0.118 0.0465 -2.54 1.12e- 2
4 mealSC 1.43 0.104 13.7 1.37e-42
5 mealUndefined 0.570 0.188 3.03 2.47e- 3step_dummy()Converts nominal data into numeric dummy variables, needed as predictors for models like KNN.
How does recipes know which variables are numeric and which are nominal?
How does recipes know what is a predictor and what is an outcome?
The formula → indicates outcomes vs predictors
The data → is only used to catalog the names and types of each variable
Helper functions for selecting sets of variables
| selector | description |
|---|---|
|
Each x variable (right side of ~) |
|
Each y variable (left side of ~) |
|
Each numeric variable |
|
Each categorical variable (e.g. factor, string) |
|
Each categorical variable (e.g. factor, string) that is defined as a predictor |
|
Each numeric variable that is defined as a predictor |
|
|
What would happen if you try to normalize a variable that doesn’t vary?
Error! You’d be dividing by zero!
step_zv()Intelligently handles zero variance variables (variables that contain only a single value)
step_normalize()Centers then scales numeric variable (mean = 0, sd = 1)
step_downsample()Unscramble! You have all the steps from our knn_rec- your challenge is to unscramble them into the right order!
Save the result as knn_rec
03:00
usemodelsusemodelshttps://tidymodels.github.io/usemodels/
kknn_recipe <-
recipe(formula = children ~ ., data = hotels) %>%
## Since distance calculations are used, the predictor variables should
## be on the same scale. Before centering and scaling the numeric
## predictors, any predictors with a single unique value are filtered
## out.
step_zv(all_predictors()) %>%
step_normalize(all_numeric_predictors())
kknn_spec <-
nearest_neighbor() %>%
set_mode("classification") %>%
set_engine("kknn")
kknn_workflow <-
workflow() %>%
add_recipe(kknn_recipe) %>%
add_model(kknn_spec) glmnet_recipe <-
recipe(formula = children ~ ., data = hotels) %>%
## Regularization methods sum up functions of the model slope
## coefficients. Because of this, the predictor variables should be on
## the same scale. Before centering and scaling the numeric predictors,
## any predictors with a single unique value are filtered out.
step_zv(all_predictors()) %>%
step_normalize(all_numeric_predictors())
glmnet_spec <-
logistic_reg() %>%
set_mode("classification") %>%
set_engine("glmnet")
glmnet_workflow <-
workflow() %>%
add_recipe(glmnet_recipe) %>%
add_model(glmnet_spec) Note
usemodels creates boilerplate code using the older pipe operator %>%
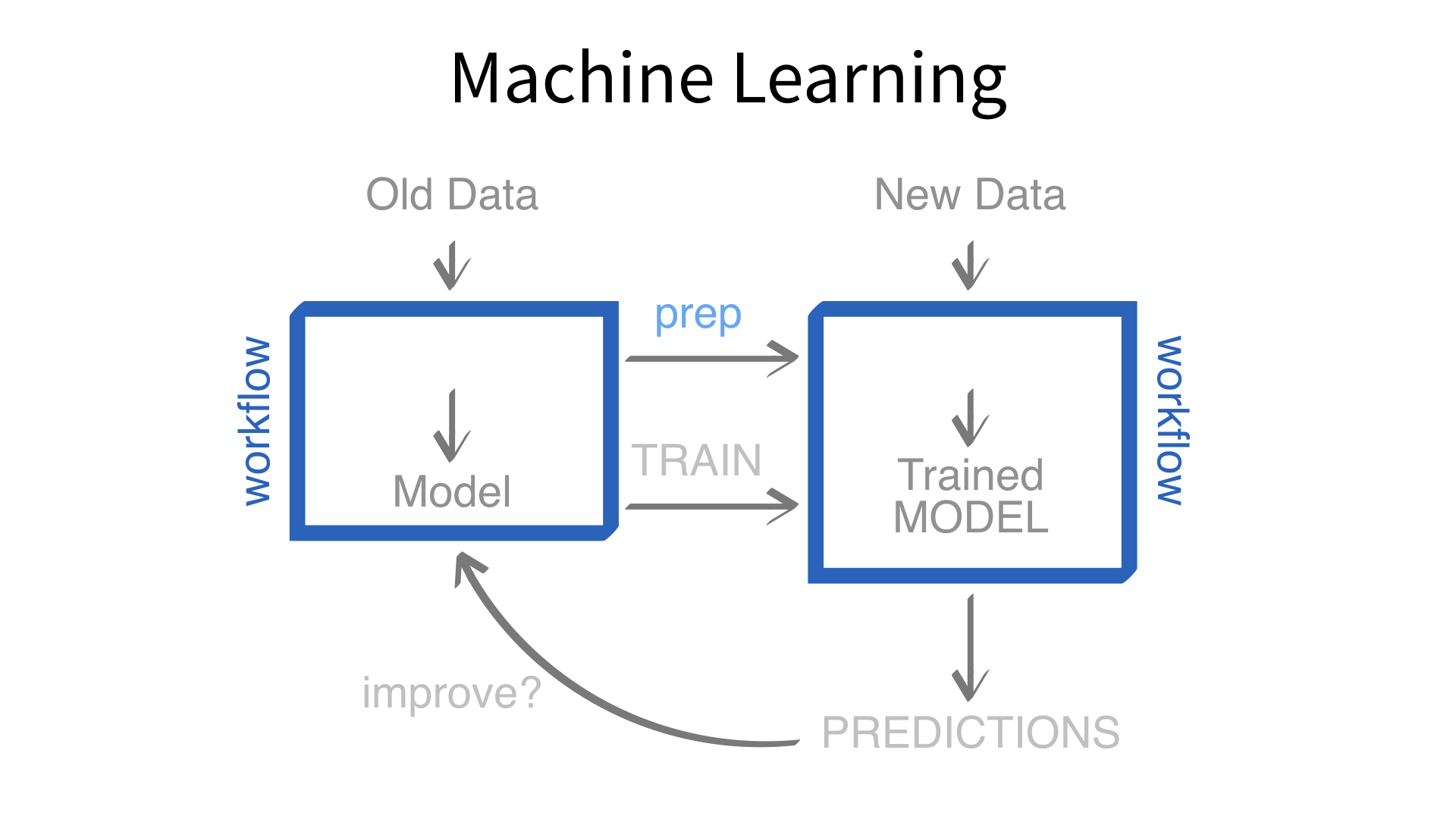
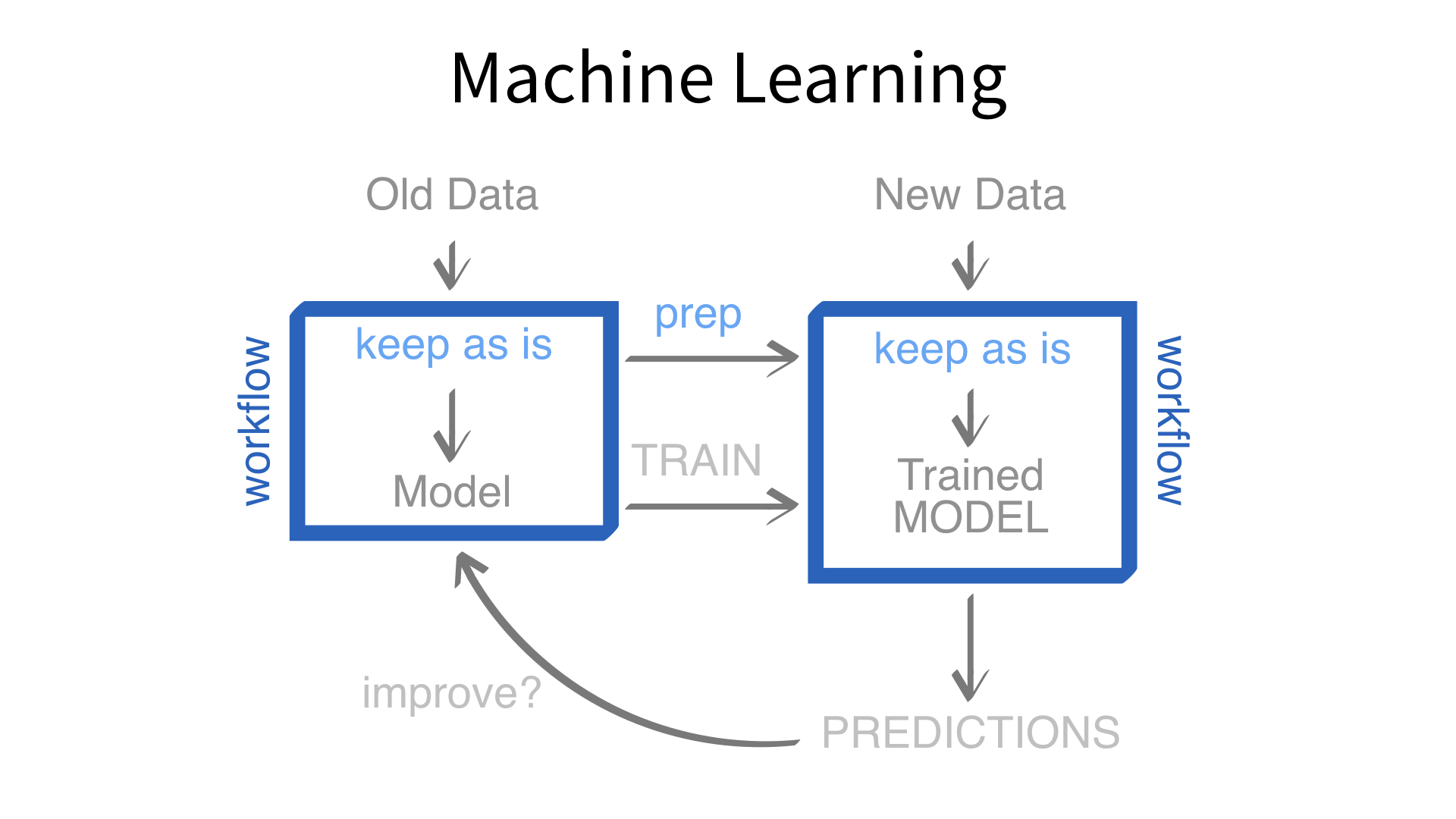
Now we’ve built a recipe.
But, how do we use a recipe?
Feature engineering and modeling are two halves of a single predictive workflow.
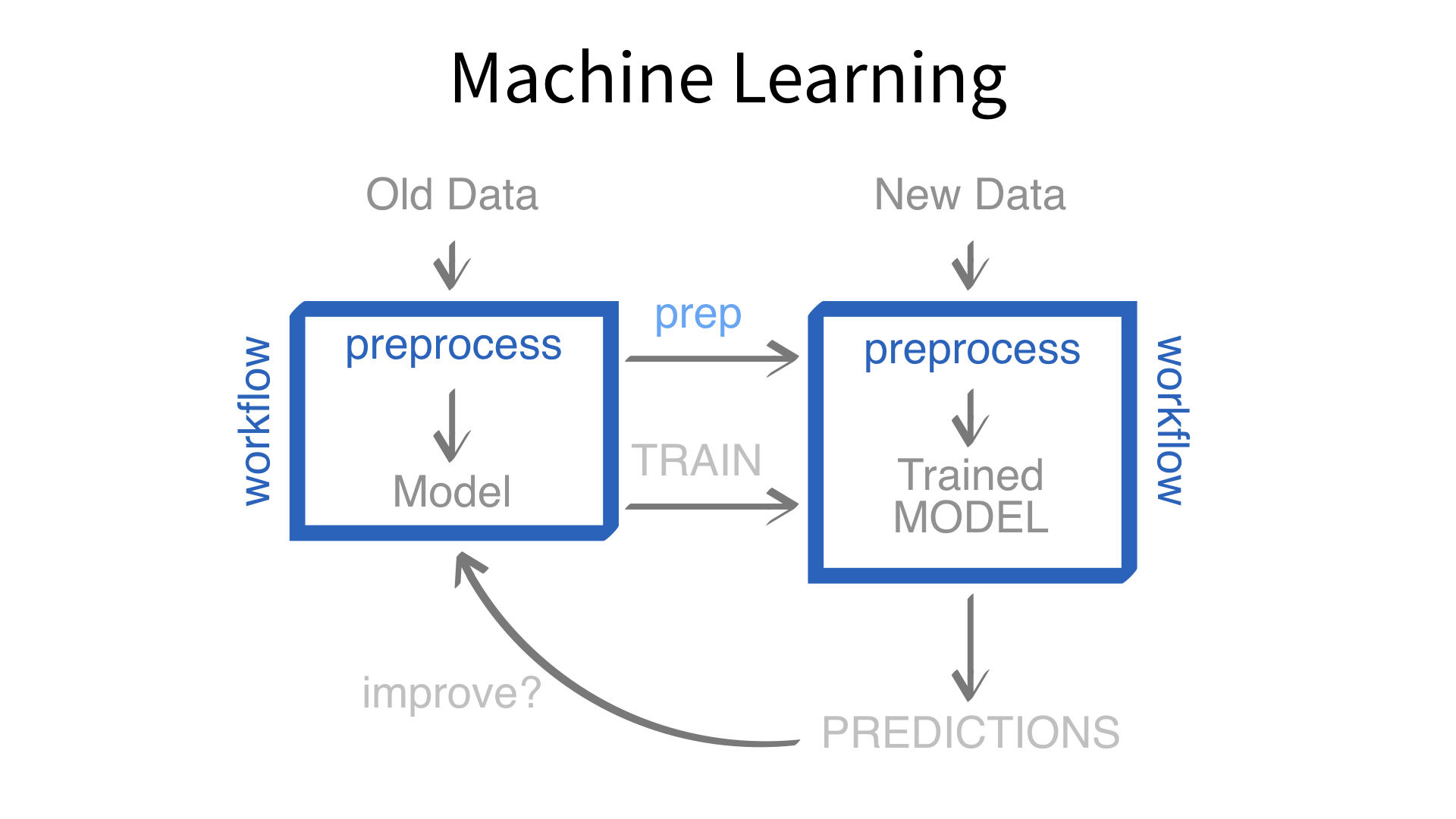
workflow()workflow()Creates a workflow to which you can add a model (and more)
add_formula()Adds a formula to a workflow *
add_model()Adds a parsnip model spec to a workflow
If we use add_model() to add a model to a workflow, what would we use to add a recipe?
Let’s see!
Fill in the blanks to make a workflow that combines knn_rec and with knn_mod.
01:00
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: nearest_neighbor()
── Preprocessor ────────────────────────────────────────────────────────────────
7 Recipe Steps
• step_date()
• step_holiday()
• step_rm()
• step_dummy()
• step_zv()
• step_normalize()
• step_downsample()
── Model ───────────────────────────────────────────────────────────────────────
K-Nearest Neighbor Model Specification (classification)
Computational engine: kknn add_recipe()Adds a recipe to a workflow.
Do you need to add a formula if you have a recipe?
fit()Fit a workflow that bundles a recipe* and a model.
fit_resamples()Fit a workflow that bundles a recipe* and a model with resampling.
Run the first chunk. Then try our KNN workflow on hotels_folds. What is the ROC AUC?
03:00
set.seed(100)
hotels_folds <- vfold_cv(hotels_train, v = 10, strata = children)
knn_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.741 10 0.00209 Preprocessor1_Mod…
2 roc_auc binary 0.830 10 0.00231 Preprocessor1_Mod…update_recipe()Replace the recipe in a workflow.
update_model()Replace the model in a workflow.
Turns out, the same knn_rec recipe can also be used to fit a penalized logistic regression model. Let’s try it out!
plr_mod <- logistic_reg(penalty = .01, mixture = 1) |>
set_engine("glmnet") |>
set_mode("classification")
plr_mod |>
translate()Logistic Regression Model Specification (classification)
Main Arguments:
penalty = 0.01
mixture = 1
Computational engine: glmnet
Model fit template:
glmnet::glmnet(x = missing_arg(), y = missing_arg(), weights = missing_arg(),
alpha = 1, family = "binomial")03:00
glmnet_wf <- knn_wf |>
update_model(plr_mod)
glmnet_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.828 10 0.00218 Preprocessor1_Mod…
2 roc_auc binary 0.873 10 0.00210 Preprocessor1_Mod…workflow() to create explicitly, logical pipelines for training a machine learning model