library(tidyverse)
library(tidymodels)
library(skimr)
library(gt) # for tables
chipotle <- read_csv(file = "data/chipotle.csv")How much food do you get when you order Chipotle?
Suggested answers
Inspired by this Reddit post, we will explore the differences in the weight of Chipotle orders. The data was originally collected by Zackary Smigel, and a cleaned copy be found in data/chipotle.csv.
Throughout the application exercise we will use the infer package which is part of tidymodels to estimate our bootstrap confidence intervals (CIs).
skim(chipotle)| Name | chipotle |
| Number of rows | 30 |
| Number of columns | 7 |
| _______________________ | |
| Column type frequency: | |
| character | 4 |
| Date | 1 |
| numeric | 2 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| order | 0 | 1 | 6 | 6 | 0 | 2 | 0 |
| meat | 0 | 1 | 7 | 8 | 0 | 2 | 0 |
| store | 0 | 1 | 7 | 7 | 0 | 3 | 0 |
| food | 0 | 1 | 4 | 7 | 0 | 2 | 0 |
Variable type: Date
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| date | 0 | 1 | 2024-01-12 | 2024-02-10 | 2024-01-26 | 30 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| day | 0 | 1 | 15.5 | 8.80 | 1.00 | 8.25 | 15.50 | 22.75 | 30.00 | ▇▇▇▇▇ |
| weight | 0 | 1 | 810.8 | 123.37 | 510.29 | 715.82 | 793.79 | 907.18 | 1048.93 | ▁▆▇▇▂ |
The variable we will use in this analysis is weight which records the total weight of the order (in grams).
Typical weight of a Chipotle order
Observed sample
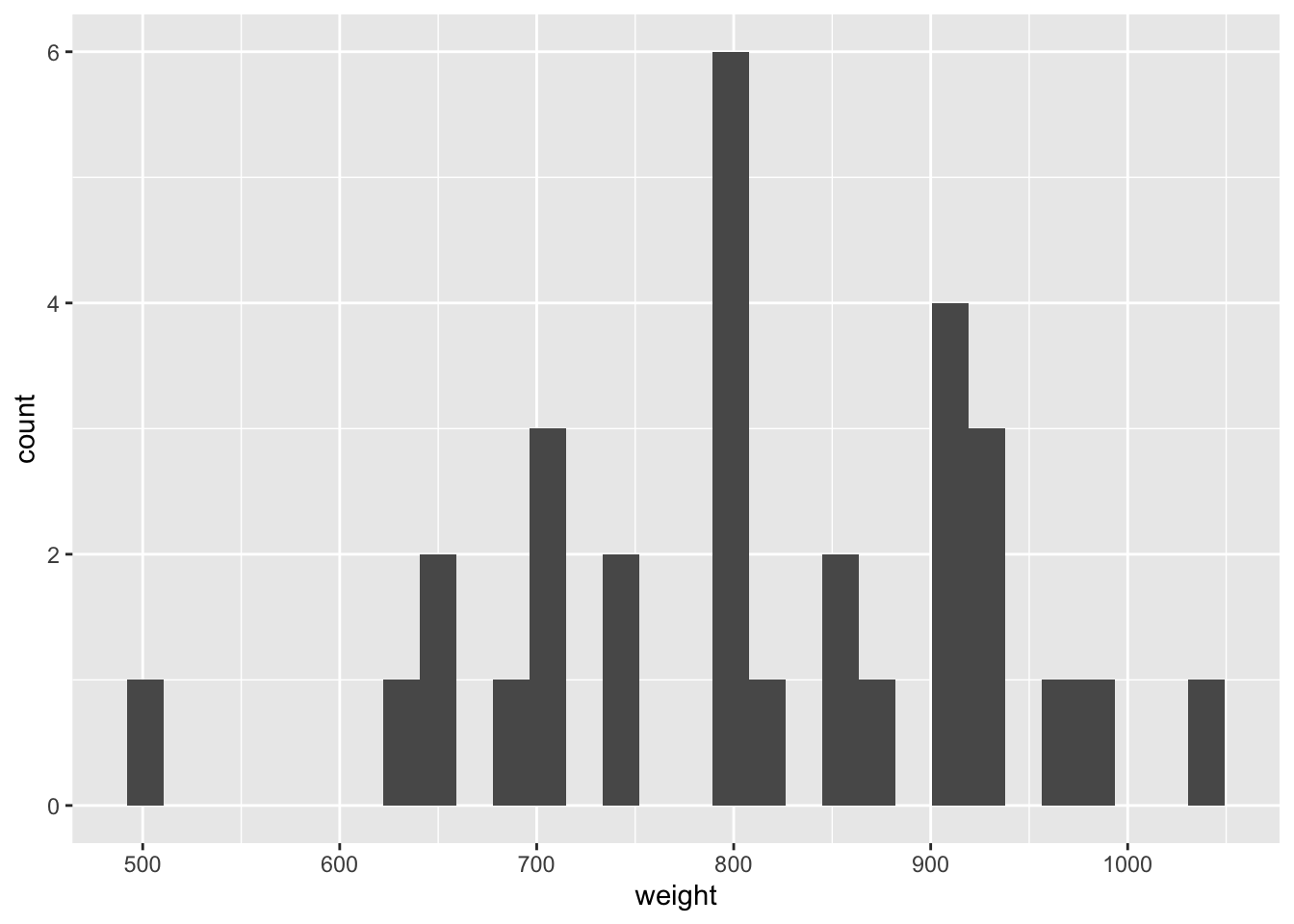
Demo: Visualize the distribution of weight using a histogram and calculate the arithmetic mean.
ggplot(data = chipotle, mapping = aes(x = weight)) +
geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.chipotle |>
summarize(mean = mean(weight))# A tibble: 1 × 1
mean
<dbl>
1 811.Your turn:
- What is the sample statistic? 810
- How large will each bootstrap sample be? 30 observations
- How many bootstrap resamples do we need? Enough
Generate bootstrap means
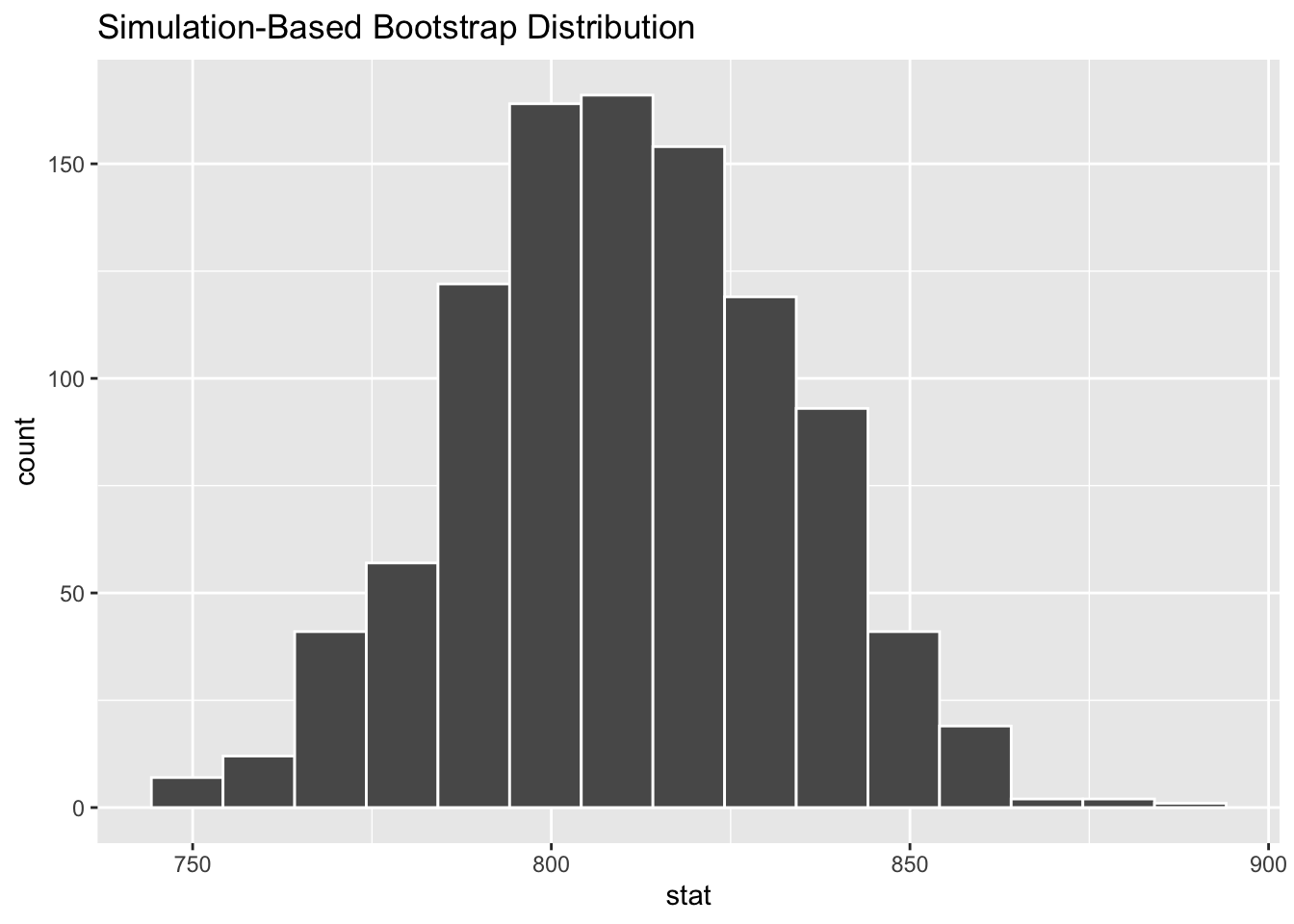
Demo: Generate 1,000 bootstrap means of the order weight.
set.seed(123) # to ensure reproducibility
# observed difference
d_hat <- chipotle |>
specify(response = weight) |>
calculate(stat = "mean")
boot_df <- chipotle |>
specify(response = weight) |>
generate(reps = 1000, type = "bootstrap") |>
calculate(stat = "mean")Your turn: Take a glimpse() of boot_df. What do you see?
glimpse(boot_df)Rows: 1,000
Columns: 2
$ replicate <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
$ stat <dbl> 786.2261, 790.0061, 803.2358, 750.3168, 773.9413, 805.1258, …Your turn: Plot a histogram of boot_df. Where is it centered? Why does this make sense?
Create a 95% confidence interval
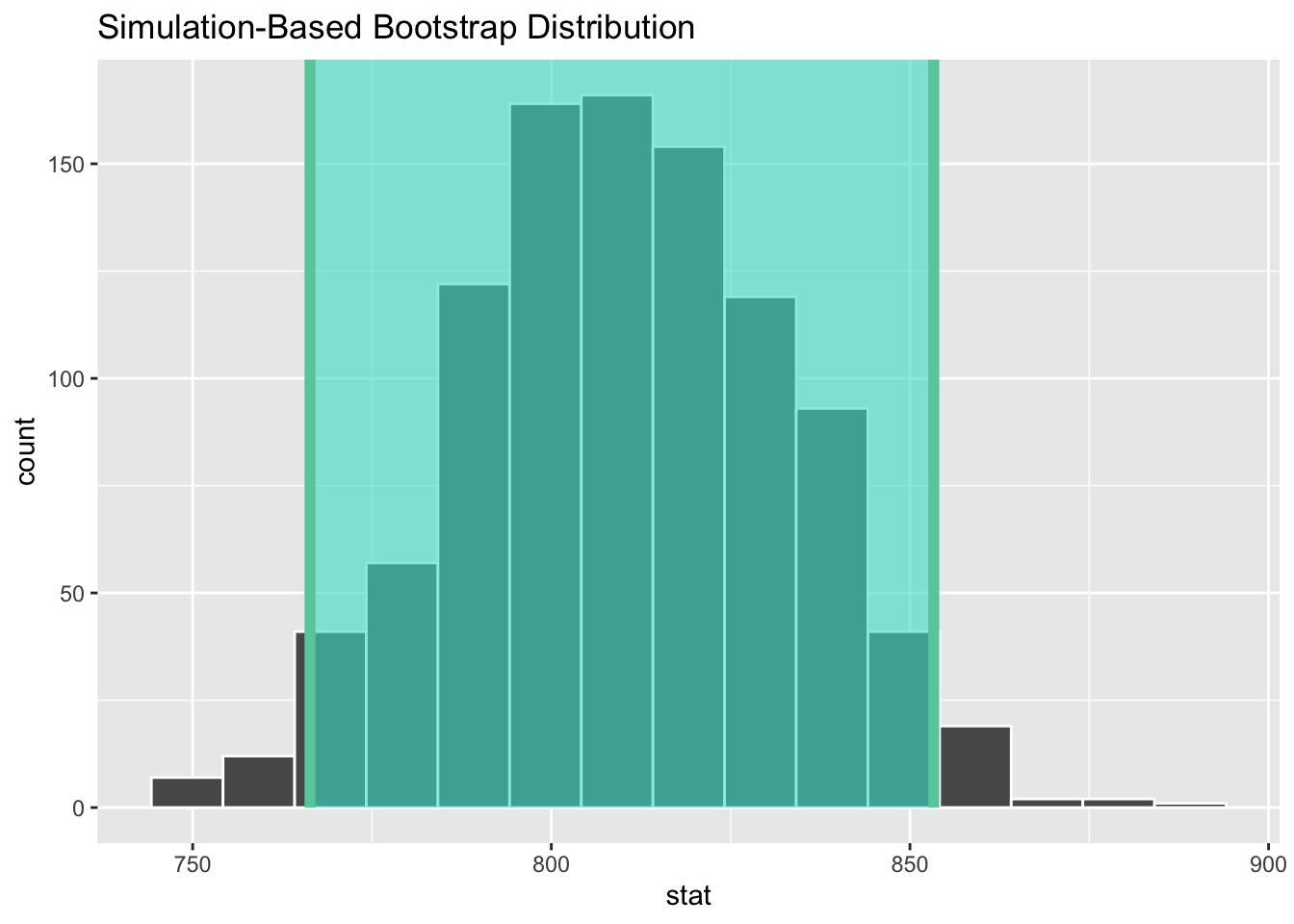
Demo: Now let’s use boot_df to create our 95% confidence interval.
get_ci(x = boot_df, level = 0.95)# A tibble: 1 × 2
lower_ci upper_ci
<dbl> <dbl>
1 766. 853.Demo: Let’s visualize our confidence interval by adding a vertical line at each of these values.
Interpreting confidence intervals
How do we interpret this confidence interval?
- There is a 95% probability the true weight of Chipotle orders is between 766 and 854 grams.
- There is a 95% probability the weight of Chipotle orders is between 766 and 854 grams.
- We are 95% confident the true weight of Chipotle orders is between 766 and 854 grams.
- We are 95% confident the weight of Chipotle orders is between 766 and 854 grams.
Alternative confidence intervals
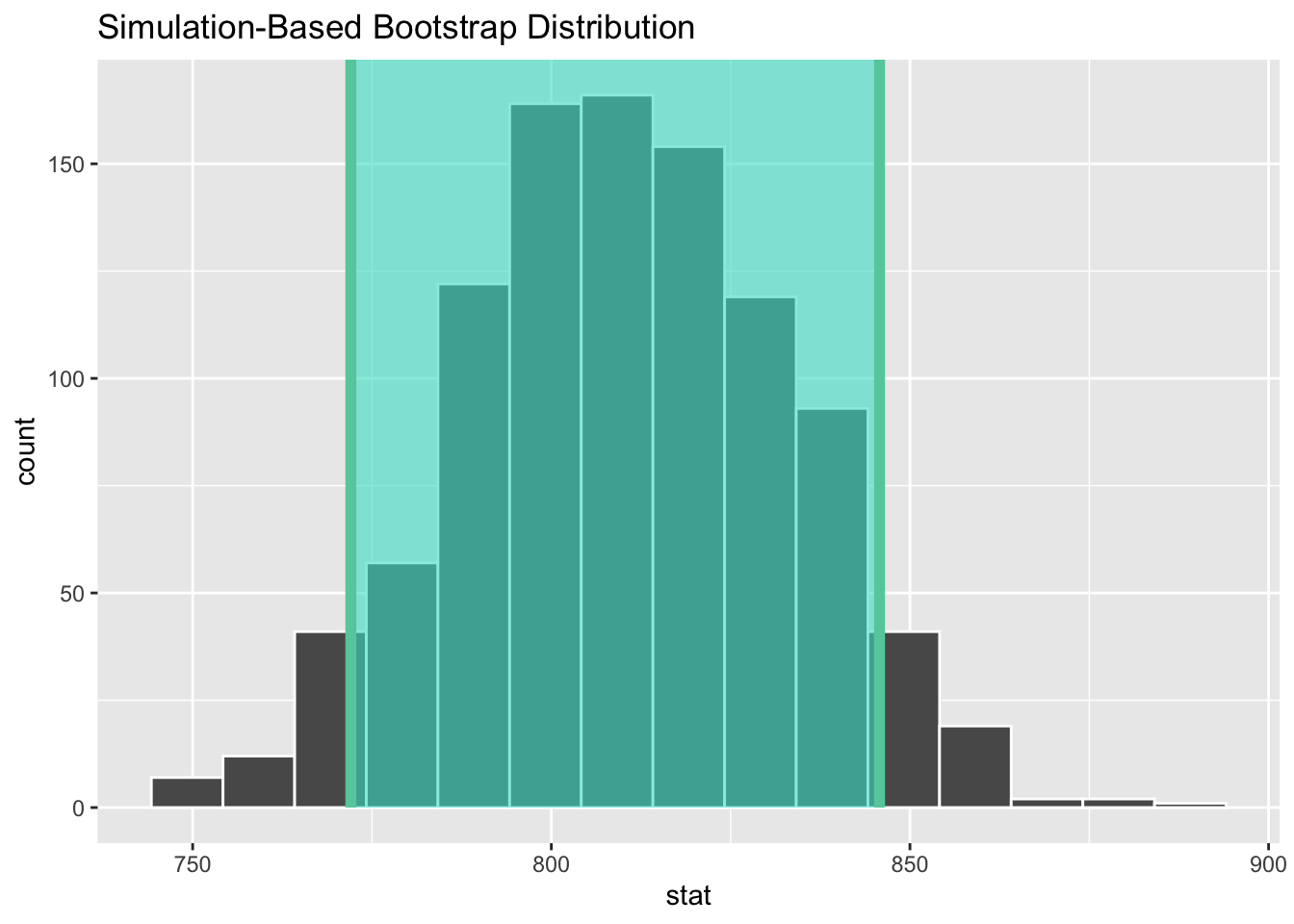
Your turn: Create a 90% confidence interval. Report it below and visualize it with the histogram created above. Is it wider or more narrow than the 95% confidence interval?
get_ci(x = boot_df, level = 0.90)# A tibble: 1 × 2
lower_ci upper_ci
<dbl> <dbl>
1 772. 846.visualize(data = boot_df) +
shade_ci(endpoints = get_ci(x = boot_df, level = 0.90))It is more narrow than the 95% confidence interval.
Difference in weight of Chipotle orders
We can calculate bootstrap confidence intervals for a range of statistical parameters.
Demo: Estimate a 95% confidence interval for the difference in means between orders placed by a person and those placed online.
# observed difference
d_hat <- chipotle |>
specify(weight ~ order) |>
calculate(stat = "diff in means", order = c("Person", "Online"))
# bootstrap CI
boot_diff_df <- chipotle |>
specify(weight ~ order) |>
generate(reps = 1000, type = "bootstrap") |>
calculate(stat = "diff in means", order = c("Person", "Online"))
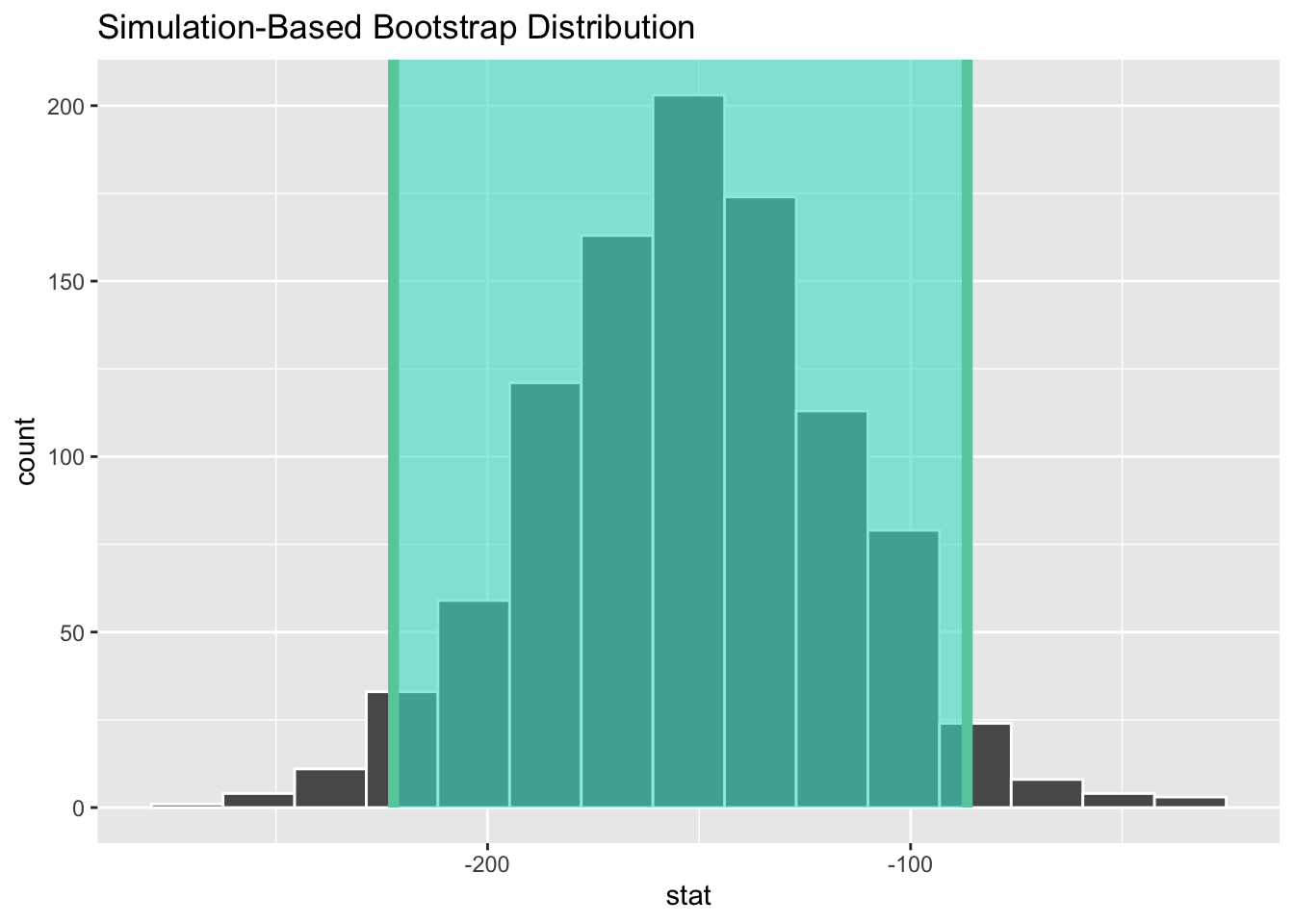
visualize(boot_diff_df) +
shade_confidence_interval(endpoints = get_ci(x = boot_diff_df, level = 0.95))get_ci(x = boot_diff_df, level = 0.95)# A tibble: 1 × 2
lower_ci upper_ci
<dbl> <dbl>
1 -222. -86.6How does that compare to the \(p\)-value from our hypothesis test?
null_dist <- chipotle |>
# specify the response and explanatory variable
specify(weight ~ order) |>
# declare the null hypothesis
hypothesize(null = "independence") |>
# simulate the null distribution
generate(reps = 1000, type = "permute") |>
# calculate the difference in means for each permutation
calculate(stat = "diff in means", order = c("Person", "Online"))
# visualize simulated p-value
visualize(null_dist) +
shade_p_value(obs_stat = d_hat, direction = "two-sided")# calculate simulated p-value
null_dist |>
get_p_value(obs_stat = d_hat, direction = "two-sided")Warning: Please be cautious in reporting a p-value of 0. This result is an
approximation based on the number of `reps` chosen in the `generate()` step.
See `?get_p_value()` for more information.# A tibble: 1 × 1
p_value
<dbl>
1 0Add response here. The \(p\)-value is less than \(0.05\), so the 95% confidence interval for the difference in means does not contain 0.
Multiple explanatory variables
We often calculate confidence intervals for the coefficients of multiple explanatory variables in a regression model. These can be useful in communicating our best guess as to the true value of the coefficient, and are often communicated as part of the results of a regression analysis.
Demo: Estimate the 95% confidence interval for the coefficients in the multiple variable model, and communicate them in a table and a coefficient plot.
# observed results
obs_fit <- chipotle |>
specify(weight ~ order + meat + food + store) |>
fit()
# null distribution for p-values
null_full_dist <- chipotle |>
specify(weight ~ order + meat + food + store) |>
hypothesize(null = "independence") |>
generate(reps = 1000, type = "permute") |>
fit()
p_vals <- get_p_value(null_full_dist, obs_stat = obs_fit, direction = "two-sided")Warning: Please be cautious in reporting a p-value of 0. This result is an
approximation based on the number of `reps` chosen in the `generate()` step.
See `?get_p_value()` for more information.# bootstrap distribution for CIs
boot_full_dist <- chipotle |>
specify(weight ~ order + meat + food + store) |>
generate(reps = 1000, type = "bootstrap") |>
fit()
# get 95% confidence interval
conf_ints <- get_ci(boot_full_dist, level = 0.95, point_estimate = obs_fit)# visualize regression results using a results table
obs_fit |>
# join with p-values
left_join(p_vals) |>
# join with confidence intervals
left_join(conf_ints) |>
# print an HTML table
gt() |>
# format for 2 significant digits
fmt_number(
columns = vars(estimate, p_value, lower_ci, upper_ci),
decimals = 2
)Joining with `by = join_by(term)`
Joining with `by = join_by(term)`Warning: Since gt v0.3.0, `columns = vars(...)` has been deprecated.
• Please use `columns = c(...)` instead.
Since gt v0.3.0, `columns = vars(...)` has been deprecated.
• Please use `columns = c(...)` instead.| term | estimate | p_value | lower_ci | upper_ci |
|---|---|---|---|---|
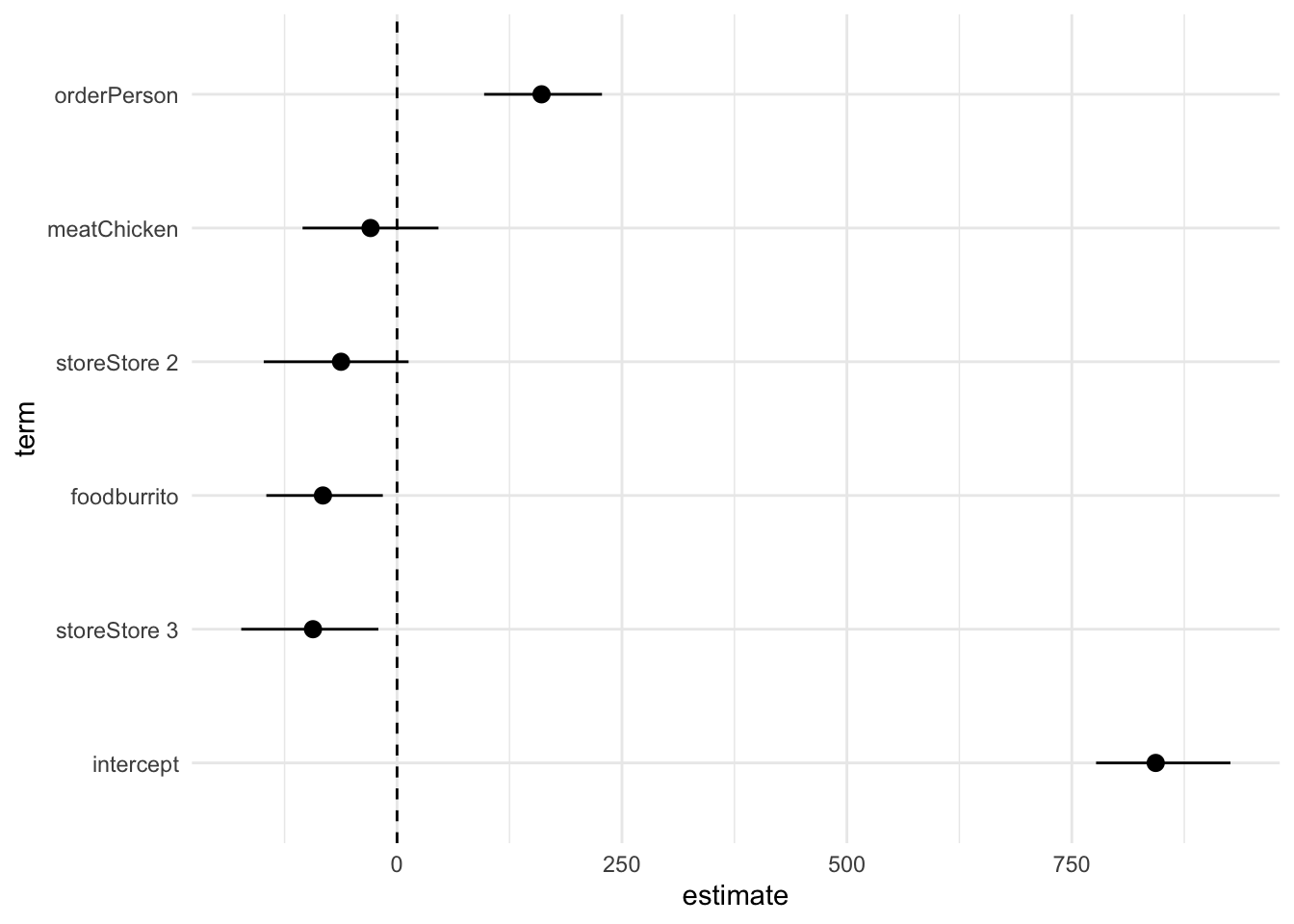
| intercept | 843.22 | 0.52 | 776.94 | 926.31 |
| orderPerson | 160.62 | 0.00 | 96.78 | 227.78 |
| meatChicken | −29.47 | 0.50 | −105.03 | 46.04 |
| foodburrito | −82.38 | 0.06 | −145.28 | −15.81 |
| storeStore 2 | −62.26 | 0.23 | −148.16 | 12.83 |
| storeStore 3 | −93.44 | 0.09 | −173.17 | −20.74 |
# visualize regression results using a coefficient plot
obs_fit |>
# join with confidence intervals
left_join(conf_ints) |>
# order the coefficients by size, pull intercept to the beginning (by convention)
mutate(term = fct_reorder(.f = term, .x = estimate) |>
fct_relevel("intercept")) |>
# draw a pointrange plot
ggplot(mapping = aes(x = estimate, y = term, xmin = lower_ci, xmax = upper_ci)) +
geom_pointrange() +
# draw a vertical line at 0
geom_vline(xintercept = 0, linetype = "dashed") +
theme_minimal()Joining with `by = join_by(term)`sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Ventura 13.6.6
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-04-13
pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.3.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
broom * 1.0.5 2023-06-09 [1] CRAN (R 4.3.0)
class 7.3-22 2023-05-03 [1] CRAN (R 4.3.2)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.2)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
data.table 1.14.10 2023-12-08 [1] CRAN (R 4.3.1)
dials * 1.2.0 2023-04-03 [1] CRAN (R 4.3.0)
DiceDesign 1.10 2023-12-07 [1] CRAN (R 4.3.1)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.1 2023-12-22 [1] CRAN (R 4.3.1)
future.apply 1.11.1 2023-12-21 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.3.1)
globals 0.16.2 2022-11-21 [1] CRAN (R 4.3.0)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gt * 0.10.0 2023-10-07 [1] CRAN (R 4.3.1)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
hardhat 1.3.0 2023-03-30 [1] CRAN (R 4.3.0)
here 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
infer * 1.0.5 2023-09-06 [1] CRAN (R 4.3.0)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
lattice 0.21-9 2023-10-01 [1] CRAN (R 4.3.2)
lava 1.7.3 2023-11-04 [1] CRAN (R 4.3.1)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
listenv 0.9.0 2022-12-16 [1] CRAN (R 4.3.0)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-60 2023-05-04 [1] CRAN (R 4.3.2)
Matrix 1.6-1.1 2023-09-18 [1] CRAN (R 4.3.2)
modeldata * 1.2.0 2023-08-09 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
nnet 7.3-19 2023-05-03 [1] CRAN (R 4.3.2)
parallelly 1.36.0 2023-05-26 [1] CRAN (R 4.3.0)
parsnip * 1.2.0 2024-02-16 [1] CRAN (R 4.3.1)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
prodlim 2023.08.28 2023-08-28 [1] CRAN (R 4.3.0)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
recipes * 1.0.9 2023-12-13 [1] CRAN (R 4.3.1)
repr 1.1.6 2023-01-26 [1] CRAN (R 4.3.0)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.1)
rpart 4.1.21 2023-10-09 [1] CRAN (R 4.3.2)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
rsample * 1.2.0 2023-08-23 [1] CRAN (R 4.3.0)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.1)
scales * 1.2.1 2024-01-18 [1] Github (r-lib/scales@c8eb772)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
skimr * 2.1.5 2022-12-23 [1] CRAN (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
survival 3.5-7 2023-08-14 [1] CRAN (R 4.3.2)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidymodels * 1.1.1 2023-08-24 [1] CRAN (R 4.3.0)
tidyr * 1.3.0 2023-01-24 [1] CRAN (R 4.3.0)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
timeDate 4032.109 2023-12-14 [1] CRAN (R 4.3.1)
tune * 1.1.2 2023-08-23 [1] CRAN (R 4.3.0)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vroom 1.6.5 2023-12-05 [1] CRAN (R 4.3.1)
withr 2.5.2 2023-10-30 [1] CRAN (R 4.3.1)
workflows * 1.1.4 2024-02-19 [1] CRAN (R 4.3.1)
workflowsets * 1.0.1 2023-04-06 [1] CRAN (R 4.3.0)
xfun 0.41 2023-11-01 [1] CRAN (R 4.3.1)
xml2 1.3.6 2023-12-04 [1] CRAN (R 4.3.1)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
yardstick * 1.3.0 2024-01-19 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────