library(tidyverse)
library(tidymodels)
library(stringr)
library(textrecipes)
library(textdata)
library(discrim)
library(themis)
library(vip)
# set seed for randomization
set.seed(123)
theme_set(theme_minimal(base_size = 13))Slay: Predicting song artist based on lyrics
Suggested answers
Import data
lyrics <- read_csv(file = "data/beyonce-swift-lyrics.csv") |>
mutate(artist = factor(artist))
lyrics# A tibble: 355 × 19
album_name track_number track_name artist lyrics danceability energy loudness
<chr> <dbl> <chr> <fct> <chr> <dbl> <dbl> <dbl>
1 COWBOY CA… 1 AMERIICAN… Beyon… "Noth… 0.374 0.515 -6.48
2 COWBOY CA… 3 16 CARRIA… Beyon… "Sixt… 0.525 0.456 -7.04
3 COWBOY CA… 5 MY ROSE Beyon… "How … 0.384 0.177 -11.8
4 COWBOY CA… 7 TEXAS HOL… Beyon… "This… 0.727 0.711 -6.55
5 COWBOY CA… 8 BODYGUARD Beyon… "One,… 0.726 0.779 -5.43
6 COWBOY CA… 10 JOLENE Beyon… "(Jol… 0.567 0.812 -5.43
7 COWBOY CA… 11 DAUGHTER Beyon… "Your… 0.374 0.448 -10.0
8 COWBOY CA… 13 ALLIIGATO… Beyon… "High… 0.618 0.651 -9.66
9 COWBOY CA… 18 FLAMENCO Beyon… "My m… 0.497 0.351 -9.25
10 COWBOY CA… 20 YA YA Beyon… "Hell… 0.617 0.904 -5.37
# ℹ 345 more rows
# ℹ 11 more variables: speechiness <dbl>, acousticness <dbl>,
# instrumentalness <dbl>, liveness <dbl>, valence <dbl>, tempo <dbl>,
# time_signature <dbl>, duration_ms <dbl>, explicit <lgl>, key_name <chr>,
# mode_name <chr>Split the data into analysis/assessment/test sets
Demonstration:
- Split the data into training/test sets with 75% allocated for training
- Split the training set into 10 cross-validation folds
# split into training/testing
set.seed(123)
lyrics_split <- initial_split(data = lyrics, strata = artist, prop = 0.75)
lyrics_train <- training(lyrics_split)
lyrics_test <- testing(lyrics_split)
# create cross-validation folds
lyrics_folds <- vfold_cv(data = lyrics_train, strata = artist)Estimate the null model for a baseline comparison
Demonstration: Estimate a null model to determine an appropriate baseline for evaluating a model’s performance.
null_spec <- null_model() |>
set_engine("parsnip") |>
set_mode("classification")
null_spec |>
fit_resamples(
# pick something as the predictor - doesn't really matter
artist ~ danceability,
resamples = lyrics_folds
) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.668 10 0.00236 Preprocessor1_Model1
2 brier_class binary 0.222 10 0.000792 Preprocessor1_Model1
3 roc_auc binary 0.5 10 0 Preprocessor1_Model1Fit a random forest model
Define the feature engineering recipe
Demonstration:
- Define a feature engineering recipe to predict the song’s artist as a function of the lyrics + audio features
- Exclude the ID variables from the recipe
- Tokenize the song lyrics
- Remove stop words
- Only keep the 500 most frequently appearing tokens
- Calculate tf-idf scores for the remaining tokens
This will generate one column for every token. Each column will have the standardized name
tfidf_lyrics_*where*is the specific token. Instead we would prefer the column names simply be*. You can remove thetfidf_lyrics_prefix using# Simplify these names step_rename_at(starts_with("tfidf_lyrics_"), fn = \(x) str_replace_all( string = x, pattern = "tfidf_lyrics_", replacement = "" ) )This does cause a conflict between the
energyaudio feature and the tokenenergy. Before removing the"tfidf_lyrics_"prefix, we will add a prefix to the audio features to avoid this conflict.# Simplify these names step_rename_at( all_predictors(), -starts_with("tfidf_lyrics_"), fn = \(x) str_glue("af_{x}") )
- Downsample the observations so there are an equal number of songs by Beyoncé and Taylor Swift in the analysis set
# define preprocessing recipe
rf_rec <- recipe(artist ~ ., data = lyrics_train) |>
# exclude ID variables
update_role(album_name, track_number, track_name, new_role = "id vars") |>
step_tokenize(lyrics) |>
step_stopwords(lyrics) |>
step_tokenfilter(lyrics, max_tokens = 500) |>
step_tfidf(lyrics) |>
# Simplify these names
step_rename_at(
all_predictors(), -starts_with("tfidf_lyrics_"),
fn = \(x) str_glue("af_{x}")
) |>
step_rename_at(starts_with("tfidf_lyrics_"),
fn = \(x) str_replace_all(
string = x,
pattern = "tfidf_lyrics_",
replacement = ""
)
) |>
step_downsample(artist)
rf_recFit the model
Demonstration:
- Define a random forest model grown with 1000 trees using the
rangerengine. - Define a workflow using the feature engineering recipe and random forest model specification. Fit the workflow using the cross-validation folds.
- Use
control = control_resamples(save_pred = TRUE)to save the assessment set predictions. We need these to assess the model’s performance. - Use
control = control_resamples(save_workflow = TRUE)to save the workflow object. We need this later on if we want to fit a single model using the same workflow and the entire training set.
- Use
# define the model specification
rf_spec <- rand_forest(trees = 1000) |>
set_mode("classification") |>
# calculate feature importance metrics using the ranger engine
set_engine("ranger", importance = "permutation")
# define the workflow
rf_wf <- workflow() |>
add_recipe(rf_rec) |>
add_model(rf_spec)
# fit the model to each of the cross-validation folds
rf_cv <- rf_wf |>
fit_resamples(
resamples = lyrics_folds,
control = control_resamples(save_pred = TRUE, save_workflow = TRUE)
)Evaluate model performance
Demonstration:
- Calculate the model’s accuracy and ROC AUC. How did it perform?
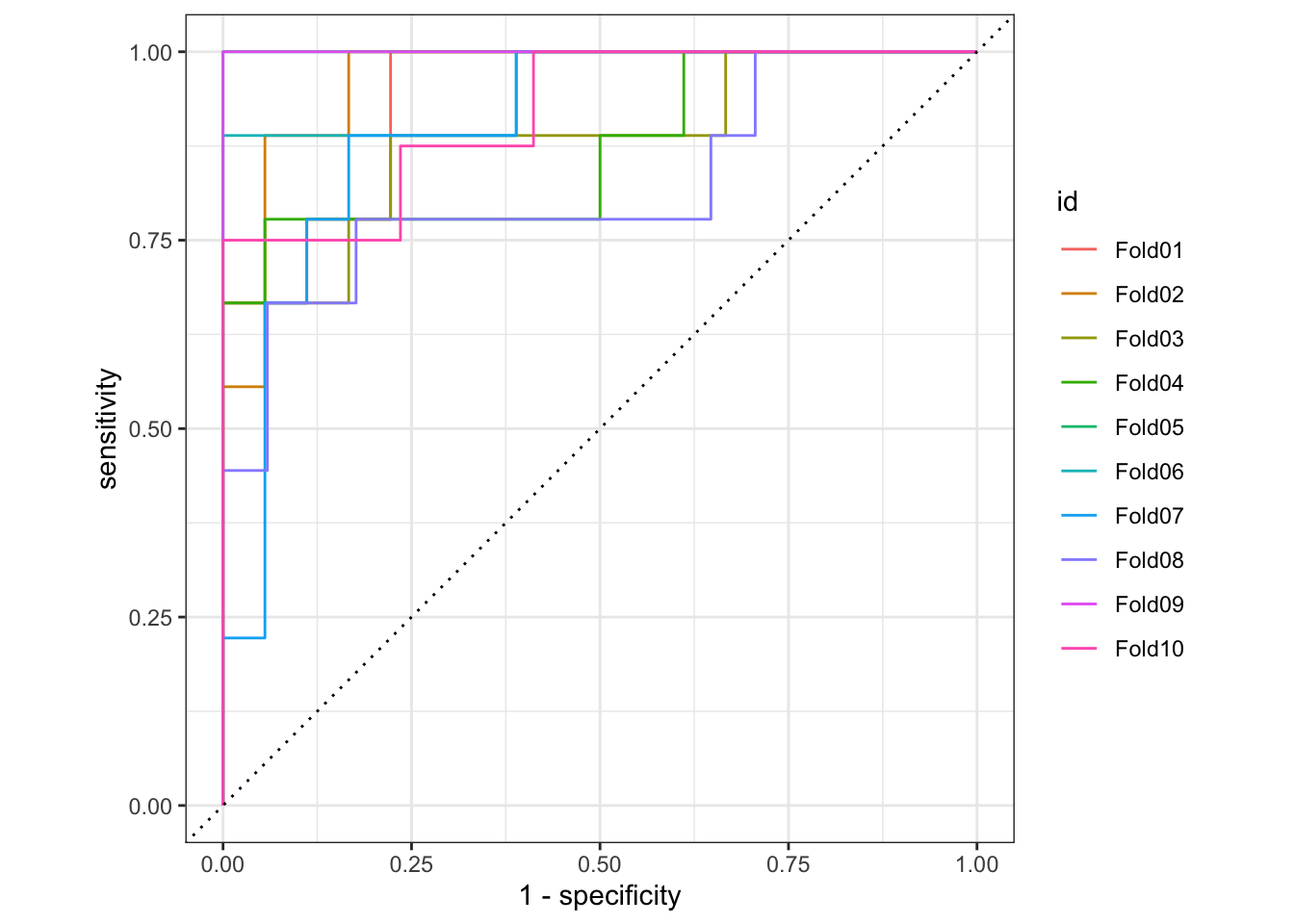
- Draw the ROC curve for each validation fold
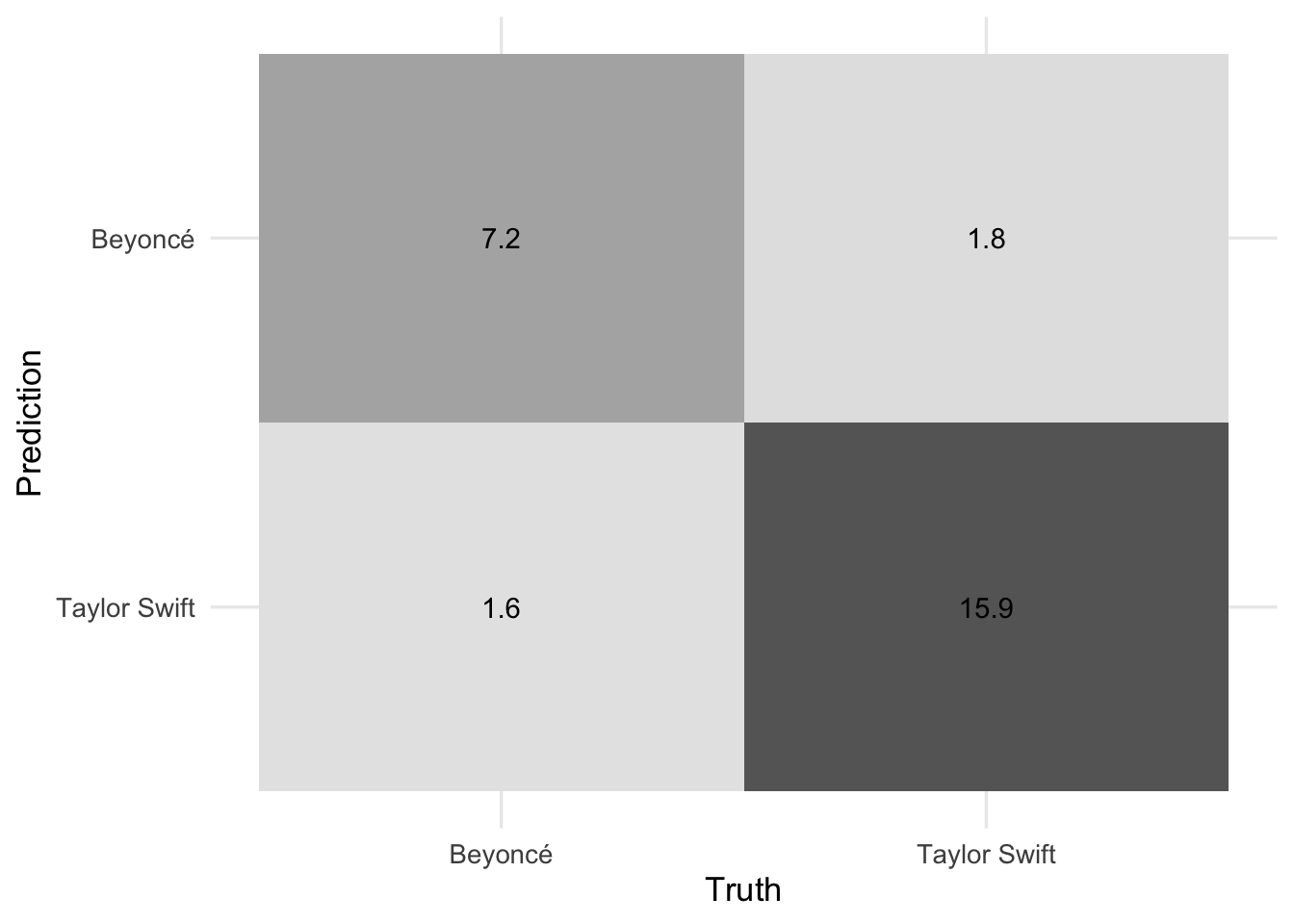
- Generate the resampled confusion matrix for the model and draw it using a heatmap. How does the model perform predicting Beyoncé songs relative to Taylor Swift songs?
# extract metrics and predictions
rf_cv_metrics <- collect_metrics(rf_cv)
rf_cv_predictions <- collect_predictions(rf_cv)
# how well did the model perform?
rf_cv_metrics# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.872 10 0.0241 Preprocessor1_Model1
2 brier_class binary 0.155 10 0.00587 Preprocessor1_Model1
3 roc_auc binary 0.925 10 0.0185 Preprocessor1_Model1# roc curve
rf_cv_predictions |>
group_by(id) |>
roc_curve(truth = artist, .pred_Beyoncé) |>
autoplot()# confusion matrix
conf_mat_resampled(x = rf_cv, tidy = FALSE) |>
autoplot(type = "heatmap")Add response here. Overall this is a pretty strong model. The ROC AUC is quite strong. The confusion matrix shows that the model is slightly better at predicting Taylor Swift songs than Beyoncé songs, but given the overrepresentation of Taylor Swift songs in the dataset this is not unusual.
Penalized regression
Define the feature engineering recipe
Demonstration:
- Define a feature engineering recipe to predict the song’s artist as a function of the lyrics + audio features
- Exclude the ID variables from the recipe
- Tokenize the song lyrics
- Calculate all possible 1-grams, 2-grams, 3-grams, 4-grams, and 5-grams
- Remove stop words
- Only keep the 2000 most frequently appearing tokens
- Calculate tf-idf scores for the remaining tokens
- Rename audio feature and tf-idf as before
- Apply required steps for penalized regression models
- Convert the
explicitvariable to a factor - Convert nominal predictors to dummy variables
- Get rid of zero-variance predictors
- Normalize all predictors to mean of 0 and variance of 1
- Convert the
- Downsample the observations so there are an equal number of songs by Beyoncé and Taylor Swift in the analysis set
glmnet_rec <- recipe(artist ~ ., data = lyrics_train) |>
# exclude ID variables
update_role(album_name, track_number, track_name, new_role = "id vars") |>
# tokenize and prep lyrics
step_tokenize(lyrics) |>
step_stopwords(lyrics) |>
step_ngram(lyrics, num_tokens = 5L, min_num_tokens = 1L) |>
step_tokenfilter(lyrics, max_tokens = 2000) |>
step_tfidf(lyrics) |>
# Simplify these names
step_rename_at(
all_predictors(), -starts_with("tfidf_lyrics_"),
fn = \(x) str_glue("af_{x}")
) |>
step_rename_at(starts_with("tfidf_lyrics_"),
fn = \(x) str_replace_all(
string = x,
pattern = "tfidf_lyrics_",
replacement = ""
)
) |>
# fix explicit variable to factor
step_bin2factor(af_explicit) |>
# normalize for penalized regression
step_dummy(all_nominal_predictors()) |>
step_zv(all_predictors()) |>
step_normalize(all_numeric_predictors()) |>
step_downsample(artist)
glmnet_recTune the penalized regression model
Demonstration:
- Define the penalized regression model specification, including tuning placeholders for
penaltyandmixture - Create the workflow object
- Define a tuning grid with every combination of:
penalty = 10^seq(-6, -1, length.out = 20)mixture = c(0, 0.2, 0.4, 0.6, 0.8, 1)
- Tune the model using the cross-validation folds
- Evaluate the tuning procedure and identify the best performing models based on ROC AUC
# define the penalized regression model specification
glmnet_spec <- logistic_reg(penalty = tune(), mixture = tune()) |>
set_mode("classification") |>
set_engine("glmnet")
# define the new workflow
glmnet_wf <- workflow() |>
add_recipe(glmnet_rec) |>
add_model(glmnet_spec)
# create the tuning grid
glmnet_grid <- expand_grid(
penalty = 10^seq(-6, -1, length.out = 20),
mixture = c(0, 0.2, 0.4, 0.6, 0.8, 1)
)
# tune over the model hyperparameters
glmnet_tune <- tune_grid(
object = glmnet_wf,
resamples = lyrics_folds,
grid = glmnet_grid,
control = control_grid(save_pred = TRUE, save_workflow = TRUE)
)# evaluate results
collect_metrics(x = glmnet_tune)# A tibble: 360 × 8
penalty mixture .metric .estimator mean n std_err .config
<dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.000001 0 accuracy binary 0.845 10 0.0186 Preprocessor1_…
2 0.000001 0 brier_class binary 0.156 10 0.00635 Preprocessor1_…
3 0.000001 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_…
4 0.00000183 0 accuracy binary 0.845 10 0.0186 Preprocessor1_…
5 0.00000183 0 brier_class binary 0.156 10 0.00635 Preprocessor1_…
6 0.00000183 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_…
7 0.00000336 0 accuracy binary 0.845 10 0.0186 Preprocessor1_…
8 0.00000336 0 brier_class binary 0.156 10 0.00635 Preprocessor1_…
9 0.00000336 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_…
10 0.00000616 0 accuracy binary 0.845 10 0.0186 Preprocessor1_…
# ℹ 350 more rowsautoplot(glmnet_tune)# identify the five best hyperparameter combinations
show_best(x = glmnet_tune, metric = "roc_auc")# A tibble: 5 × 8
penalty mixture .metric .estimator mean n std_err .config
<dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.000001 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_Model…
2 0.00000183 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_Model…
3 0.00000336 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_Model…
4 0.00000616 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_Model…
5 0.0000113 0 roc_auc binary 0.881 10 0.0249 Preprocessor1_Model…Add response here. The best performing hyperparameter combination has a strong ROC AUC value, though it is slightly lower compared to the random forest model.
Random forest with word embeddings
Define the feature engineering recipe
Demonstration: Rather than using individual tokens and tf-idf scores, we will use pre-trained word embeddings to represent the lyrics. We will calculate the aggregate embedding for each song by averaging the embeddings of all the words in the song, then tune a random forest model using these embeddings.
Normally in order to import GloVe embeddings you would use the code below:
glove_embed <- embedding_glove6b(dimensions = 100)This downloads the ZIP file containing the embeddings, stores it in a cache folder, and then imports the requested embeddings and dimensions as a data frame. Note that many of the embeddings are stored in ZIP files that are multiple gigabytes in size. Often it is easier to manually download the files and store them in the appropriate location outside of R. See the documentation for embedding_glove*() for more information.
# hacky way to make it work on RStudio Workbench
glove_embed <- read_delim(
file = "/rstudio-files/glove6b/glove.6B.100d.txt",
delim = " ",
quote = "",
col_names = c(
"token",
paste0("d", seq_len(100))
),
col_types = paste0(
c(
"c",
rep("d", 100)
),
collapse = ""
)
)# load previously-downloaded word embeddings
glove_embed <- embedding_glove6b(
# dir = "/rstudio-files",
dimensions = 100,
manual_download = TRUE
)rf_embeds_rec <- recipe(artist ~ ., data = lyrics_train) |>
# exclude ID variables
update_role(album_name, track_number, track_name, new_role = "id vars") |>
step_tokenize(lyrics) |>
# calculate aggregate embedding for each song
step_word_embeddings(lyrics, embeddings = glove_embed, aggregation = "mean") |>
step_downsample(artist)
rf_embeds_recTune the model
Demonstration:
- Define the penalized regression model specification, including tuning placeholders for
mtryandmin_n - Create the workflow object
- Tune the model using the cross-validation folds and an automatically generated tuning grid
- Evaluate the tuning procedure and identify the best performing models based on ROC AUC
# define the model specification
rf_embeds_spec <- rand_forest(trees = 1000, mtry = tune(), min_n = tune()) |>
set_mode("classification") |>
# calculate feature importance metrics using the ranger engine
set_engine("ranger", importance = "permutation")
# define the workflow
rf_embeds_wf <- workflow() |>
add_recipe(rf_embeds_rec) |>
add_model(rf_embeds_spec)
# fit the model to each of the cross-validation folds
rf_embeds_cv <- rf_embeds_wf |>
tune_grid(
resamples = lyrics_folds,
control = control_resamples(save_pred = TRUE, save_workflow = TRUE)
)# extract metrics
rf_embeds_cv_metrics <- collect_metrics(rf_embeds_cv)
# how well did the model perform?
rf_embeds_cv_metrics# A tibble: 30 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 104 35 accuracy binary 0.817 10 0.0278 Preprocessor1_Model01
2 104 35 brier_class binary 0.145 10 0.00821 Preprocessor1_Model01
3 104 35 roc_auc binary 0.889 10 0.0231 Preprocessor1_Model01
4 58 38 accuracy binary 0.809 10 0.0278 Preprocessor1_Model02
5 58 38 brier_class binary 0.146 10 0.00793 Preprocessor1_Model02
6 58 38 roc_auc binary 0.894 10 0.0213 Preprocessor1_Model02
7 93 28 accuracy binary 0.828 10 0.0309 Preprocessor1_Model03
8 93 28 brier_class binary 0.143 10 0.00820 Preprocessor1_Model03
9 93 28 roc_auc binary 0.900 10 0.0204 Preprocessor1_Model03
10 45 24 accuracy binary 0.832 10 0.0273 Preprocessor1_Model04
# ℹ 20 more rowsshow_best(rf_embeds_cv, metric = "roc_auc")# A tibble: 5 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 78 16 roc_auc binary 0.902 10 0.0216 Preprocessor1_Model05
2 93 28 roc_auc binary 0.900 10 0.0204 Preprocessor1_Model03
3 26 8 roc_auc binary 0.900 10 0.0220 Preprocessor1_Model09
4 18 13 roc_auc binary 0.900 10 0.0204 Preprocessor1_Model10
5 90 20 roc_auc binary 0.896 10 0.0200 Preprocessor1_Model08Add response here. The random forest model using word embeddings performed better than the penalized regression model, but not as well as the random forest model using tf-idf scores. That said, all the ROC AUC values are quite close to each other.
Fit the best model
Your turn:
- Select the model + hyperparameter combinations that achieve the highest ROC AUC
- Fit that model using the best hyperparameters and the full training set. How well does the model perform on the test set?
# select the best model's hyperparameters
best_fit <- fit_best(rf_cv)
# test set ROC AUC
bind_cols(
lyrics_test,
predict(best_fit, new_data = lyrics_test, type = "prob")
) |>
roc_auc(truth = artist, .pred_Beyoncé)# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.942Add response here. The test set ROC AUC is slightly higher than the cross-validated metrics, indicating that the model generalizes well to new data.
Variable importance
We can examine the results of each model to evaluate which tokens were the most important in generating artist predictions. Here we use vip to calculate importance.
# extract parsnip model fit
rf_imp <- rf_cv |>
fit_best() |>
extract_fit_parsnip() |>
vi(method = "model")
# clean up the data frame for visualization
rf_imp |>
# extract 20 most important n-grams
slice_max(order_by = Importance, n = 20) |>
mutate(Variable = fct_reorder(.f = Variable, .x = Importance)) |>
ggplot(mapping = aes(
x = Importance,
y = Variable
)) +
geom_col() +
labs(
y = NULL,
title = "Most relevant features for predicting whether\na song is by Beyoncé or Taylor Swift",
subtitle = "Random forest model"
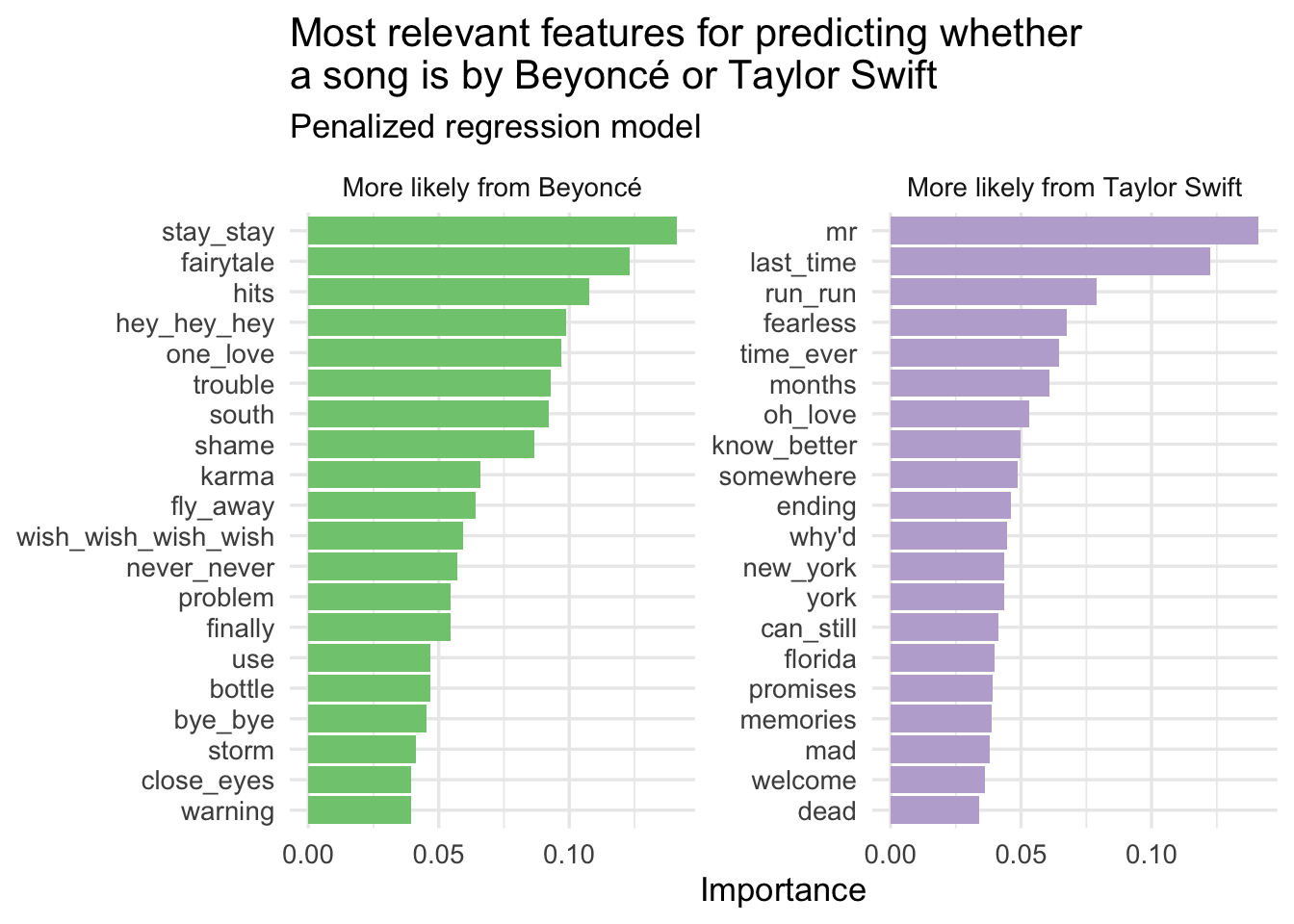
)# extract parsnip model fit
glmnet_imp <- glmnet_tune |>
fit_best() |>
extract_fit_parsnip() |>
vi(method = "model", lambda = select_best(x = glmnet_tune, metric = "roc_auc")$penalty)
# clean up the data frame for visualization
glmnet_imp |>
mutate(
Sign = case_when(
Sign == "NEG" ~ "More likely from Beyoncé",
Sign == "POS" ~ "More likely from Taylor Swift"
),
Importance = abs(Importance)
) |>
# importance must be greater than 0
filter(Importance > 0) |>
# keep top 20 features for each artist
slice_max(n = 20, order_by = Importance, by = Sign) |>
mutate(Variable = fct_reorder(.f = Variable, .x = Importance)) |>
ggplot(mapping = aes(
x = Importance,
y = Variable,
fill = Sign
)) +
geom_col(show.legend = FALSE) +
scale_fill_brewer(type = "qual") +
facet_wrap(facets = vars(Sign), scales = "free_y") +
labs(
y = NULL,
title = "Most relevant features for predicting whether\na song is by Beyoncé or Taylor Swift",
subtitle = "Penalized regression model"
)sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Ventura 13.6.6
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-05-06
pandoc 3.1.11 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/aarch64/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
archive 1.1.7 2023-12-11 [1] CRAN (R 4.3.1)
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
broom * 1.0.5 2023-06-09 [1] CRAN (R 4.3.0)
class 7.3-22 2023-05-03 [1] CRAN (R 4.3.2)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
codetools 0.2-19 2023-02-01 [1] CRAN (R 4.3.2)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
data.table 1.15.4 2024-03-30 [1] CRAN (R 4.3.1)
dials * 1.2.1 2024-02-22 [1] CRAN (R 4.3.1)
DiceDesign 1.10 2023-12-07 [1] CRAN (R 4.3.1)
digest 0.6.35 2024-03-11 [1] CRAN (R 4.3.1)
discrim * 1.0.1 2023-03-08 [1] CRAN (R 4.3.0)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
foreach 1.5.2 2022-02-02 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
furrr 0.3.1 2022-08-15 [1] CRAN (R 4.3.0)
future 1.33.2 2024-03-26 [1] CRAN (R 4.3.1)
future.apply 1.11.2 2024-03-28 [1] CRAN (R 4.3.1)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.3.1)
glmnet * 4.1-8 2023-08-22 [1] CRAN (R 4.3.0)
globals 0.16.3 2024-03-08 [1] CRAN (R 4.3.1)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
gower 1.0.1 2022-12-22 [1] CRAN (R 4.3.0)
GPfit 1.0-8 2019-02-08 [1] CRAN (R 4.3.0)
gtable 0.3.5 2024-04-22 [1] CRAN (R 4.3.1)
hardhat 1.3.1 2024-02-02 [1] CRAN (R 4.3.1)
here 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
infer * 1.0.7 2024-03-25 [1] CRAN (R 4.3.1)
ipred 0.9-14 2023-03-09 [1] CRAN (R 4.3.0)
iterators 1.0.14 2022-02-05 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
lattice 0.21-9 2023-10-01 [1] CRAN (R 4.3.2)
lava 1.8.0 2024-03-05 [1] CRAN (R 4.3.1)
lhs 1.1.6 2022-12-17 [1] CRAN (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
listenv 0.9.1 2024-01-29 [1] CRAN (R 4.3.1)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
MASS 7.3-60 2023-05-04 [1] CRAN (R 4.3.2)
Matrix * 1.6-1.1 2023-09-18 [1] CRAN (R 4.3.2)
modeldata * 1.3.0 2024-01-21 [1] CRAN (R 4.3.1)
modelenv 0.1.1 2023-03-08 [1] CRAN (R 4.3.0)
munsell 0.5.1 2024-04-01 [1] CRAN (R 4.3.1)
nnet 7.3-19 2023-05-03 [1] CRAN (R 4.3.2)
parallelly 1.37.1 2024-02-29 [1] CRAN (R 4.3.1)
parsnip * 1.2.1 2024-03-22 [1] CRAN (R 4.3.1)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
prodlim 2023.08.28 2023-08-28 [1] CRAN (R 4.3.0)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
ranger * 0.16.0 2023-11-12 [1] CRAN (R 4.3.1)
rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.3.0)
RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
recipes * 1.0.10 2024-02-18 [1] CRAN (R 4.3.1)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.26 2024-03-05 [1] CRAN (R 4.3.1)
ROSE 0.0-4 2021-06-14 [1] CRAN (R 4.3.0)
rpart 4.1.21 2023-10-09 [1] CRAN (R 4.3.2)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
rsample * 1.2.1 2024-03-25 [1] CRAN (R 4.3.1)
rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.3.1)
scales * 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
shape 1.4.6.1 2024-02-23 [1] CRAN (R 4.3.1)
SnowballC 0.7.1 2023-04-25 [1] CRAN (R 4.3.0)
stopwords * 2.3 2021-10-28 [1] CRAN (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
survival 3.5-7 2023-08-14 [1] CRAN (R 4.3.2)
textdata * 0.4.4 2022-09-02 [1] CRAN (R 4.3.0)
textrecipes * 1.0.6 2023-11-15 [1] CRAN (R 4.3.1)
themis * 1.0.2 2023-08-14 [1] CRAN (R 4.3.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidymodels * 1.2.0 2024-03-25 [1] CRAN (R 4.3.1)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.3.1)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.1)
timeDate 4032.109 2023-12-14 [1] CRAN (R 4.3.1)
tokenizers 0.3.0 2022-12-22 [1] CRAN (R 4.3.0)
tune * 1.2.1 2024-04-18 [1] CRAN (R 4.3.1)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vip * 0.4.1 2023-08-21 [1] CRAN (R 4.3.0)
vroom 1.6.5 2023-12-05 [1] CRAN (R 4.3.1)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
workflows * 1.1.4 2024-02-19 [1] CRAN (R 4.3.1)
workflowsets * 1.1.0 2024-03-21 [1] CRAN (R 4.3.1)
xfun 0.43 2024-03-25 [1] CRAN (R 4.3.1)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
yardstick * 1.3.1 2024-03-21 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────